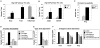Jmjd3 and UTX play a demethylase-independent role in chromatin remodeling to regulate T-box family member-dependent gene expression - PubMed (original) (raw)
Jmjd3 and UTX play a demethylase-independent role in chromatin remodeling to regulate T-box family member-dependent gene expression
Sara A Miller et al. Mol Cell. 2010.
Abstract
The stable and heritable H3K27-methyl mark suppresses transcription of lineage-specific genes in progenitor cells. During developmental transitions, histone demethylases are required to dramatically alter epigenetic and gene expression states to create new cell-specific profiles. It is unclear why demethylase proteins that antagonize polycomb-mediated repression continue to be expressed in terminally differentiated cells where further changes in H3K27 methylation could be deleterious. In this study, we show that the H3K27 demethylases, Jmjd3 and UTX, mediate a functional interaction between the lineage-defining T-box transcription factor family and a Brg1-containing SWI/SNF remodeling complex. Importantly, Jmjd3 is required for the coprecipitation of Brg1 with the T-box factor, T-bet, and this interaction is necessary for Ifng remodeling in differentiated Th1 cells. Thus, Jmjd3 has a required role in general chromatin remodeling that is independent from its H3K27 demethylase potential. This function for H3K27 demethylase proteins may explain their presence in differentiated cells where the epigenetic profile is already established.
Copyright © 2010 Elsevier Inc. All rights reserved.
Figures
Figure 1
Jmjd3 is required for T-box protein-dependent gene expression, but its H3K27-demethylase activity is not required to induce a subset of target genes. (A–C) EL4 T cells were transfected with a vector control, or T-box factor expression constructs for Eomes, Tbx5 or Tbx6 in combination with either a control siRNA (siGFP) or an siRNA to Jmjd3 (siJmjd3). (D) EL4 cells were transfected with a vector control or T-bet expression construct with either siGFP, siJmjd3, siJmjd3 and wild-type Jmjd3 or siJmjd3 and an H3K27-demethylase catalytically dead mutant of Jmjd3. (A–D) Cell aliquots were harvested for RNA and qRT-PCR and the data represent the mean of independent experiments with standard deviation.
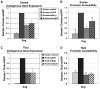
Figure 2
T-bet requires Jmjd3 for a general chromatin remodeling function that is independent from its H3K27-demethylase activity. (A–D) EL4 cells were transfected with either a vector control or a T-bet expression construct in combination with siGFP, siJmjd3 alone or (D) siJmjd3 with either a wild-type Jmjd3 or Jmjd3 catalytic mutant construct for rescue. Cell aliquots were harvested for (A) luciferase analysis, (B–D) RE assays. (A) For the luciferase analysis, the transfections were left unstimulated or treated with PMA and ionomycin to mimic TCR stimulation 6 hours post-transfection. (A–D) The data represent the mean of independent experiments and their standard deviation.
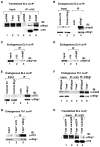
Figure 3
The T-bet-dependent recruitment of a Brg1-containing SWI/SNF complex to the Ifng promoter affects optimal promoter accessibility and endogenous gene expression similar to Jmjd3. (A–C) Primary Th1 cells from wild-type or T-bet−/− mice were analyzed in (A, B) ChIP and (C) RE assays, with the mean quantitation and standard deviation from independent experiments shown. (A, B) The ChIP assay was performed with antibodies specific for Brg1, T-bet, H3K27me3, Jmjd3, Baf170 or a non-specific IgG antibody control. Samples were normalized to an aliquot of the total input that was also amplified with gene specific primers to Ifng with the background non-specific IgG control signal subtracted. (D, E) EL4 cells were transfected with a vector control (dark grey), wild-type T-bet either with siGFP (black), siBrg1 (white spotted), siJmjd3 (light grey) or siBrg1 and siJmjd3 (grey spotted). Cells were harvested for (D) RE analysis to determine Ifng promoter accessibility or (E) qRT-PCR analysis of endogenous Ifng expression. (F) EL4 cells were transfected with a control vector or wild-type T-bet with either siGFP (black), siBaf170 (light grey), siBaf155 (spotted light grey) or siBaf170 and siBaf155 (grey with black stripes). Endogenous Ccl3, Cxcr3 and Ifng expression was monitored by qRT-PCR analysis. The mean of three independent experiments with standard deviation relative to the T-bet+siGFP sample is shown.
Figure 4
T-box factors require Jmjd3 and Brg1 for chromatin remodeling and gene expression at the Ifng promoter. EL4 cells were transfected with a vector control (grey) or T-box expression constructs for Eomes (dark grey) or Tbx5 (black spotted) with either siGFP, siBrg1 (thin stripe) or siJmjd3 (wide stripe). Samples were collected for (A, C) RNA and qRT-PCR of endogenous Ifng expression or (B, D) RE analysis to determine promoter accessibility. (A–D) Data represent the mean of independent experiments with standard deviation.
Figure 5
The functional interaction between T-bet and Jmjd3 localizes to a region that is highly homologous among H3K27-demethylase subfamily members. (A–D) EL4 cells were transfected with either a vector control or a T-bet expression construct in combination with siGFP, siJmjd3 alone or siJmjd3 with either a wild-type Jmjd3 or Jmjd3 truncation mutant constructs for rescue as indicated. The amino acids from Jmjd3 that are included within each mutant construct are indicated by the numbers in the key of each graph. (B) The three core catalytic residues were mutated within the context of the Jmjd3 1170-1483 truncation to create a catalytically dead protein. Cell aliquots were harvested for (A–C) RNA and qRT-PCR of Cxcr3 endogenous gene expression, (D) RE accessibility of the Ifng promoter, or Western analysis (Figure S6). Shown is the mean of independent experiments with standard deviation. (E) As indicated above the schematic, Jmjd3 is highly homologous to its nearest Jumonji subfamily members, UTX and UTY. The subfamily diverges in the N-terminal domain where there is only 29.1% homology (6.7% identity), but importantly are 87.4% homologous (69% identity) throughout the mixed and Jmjc domain and 77.6% (49.2% identity) in the C-terminus.
Figure 6
T-box factors also functionally interact with UTX to induce chromatin remodeling and gene expression. (A–E) EL4 cells were transfected with a vector control or T-box factor expression constructs with either siGFP, siUTX or siJmjd3 as indicated. Cell aliquots were harvested for (A–C) RNA and qRT-PCR to determine endogenous Ifng expression and (D, E) RE accessibility to examine chromatin remodeling. Shown is the mean of independent experiments with standard deviation.
Figure 7
Jmjd3 is required for T-bet to physically interact with a Brg1-containing SWI/SNF complex. Whole cell extracts from EL4 or primary Th1 T cells were prepared and immunoprecipitated with antibodies specific either to the endogenous protein or V5 epitope tag. Immunocomplexes were resolved by SDS page and Western analysis with specific antibodies as indicated to the right of the figure was performed. (A, H) EL4 T cells were transfected as indicated above each lane. The specific Brg1 band is indicated by an asterisk. As a control for the IP, the blots were reprobed with an antibody to the V5 epitope tag. Lysates from (B–E) untransfected EL4 T cells or (F,G) primary Th1 cells were precipitated with Brg1 or a control antibody as indicated above each lane. As a control, the blots were reprobed with a second Brg1 antibody. (A) Both T-bet and wild-type Jmjd3 associate with Brg1. EL4 cells were transfected with a pcDNA vector control (lanes 1, 4), T-bet (lanes 2, 5) or Jmjd3 (lanes 3, 6). (B–E) Endogenous H3K27-demethylase subfamily proteins interact with Brg1 and other components of the SWI/SNF complex. Lysates from EL4 cells were precipitated with either a control antibody or Brg1 and probed with an antibody specific to (B) Jmjd3 or (E) UTX. Additionally, EL4 lysates were precipitated with a control or Jmjd3 antibody probed with an antibody to (C) Baf170 or (D) Baf155. (F, G) H3K27-demethylase jumonji subfamily proteins form complexes with Brg1 at endogenous levels in primary Th1 cells independent of T-bet expression. (F) Lysates from wildtype and T-bet−/− Th1 cells were precipitated with a control or Brg1 antibody and blots were probed with Jmjd3. (G) Samples from T-bet−/− cells were precipitated with control or Brg1 antibody and the blot was probed for UTX. (H) Jmjd3 is required for the physical interaction between T-bet and Brg1. EL4 cells were transfected with T-bet and either a control siGFP (lanes 1, 3) or an siRNA specific to Jmjd3 (lanes 2, 4) followed by co-IP analysis.
Similar articles
- Identification of JmjC domain-containing UTX and JMJD3 as histone H3 lysine 27 demethylases.
Hong S, Cho YW, Yu LR, Yu H, Veenstra TD, Ge K. Hong S, et al. Proc Natl Acad Sci U S A. 2007 Nov 20;104(47):18439-44. doi: 10.1073/pnas.0707292104. Epub 2007 Nov 14. Proc Natl Acad Sci U S A. 2007. PMID: 18003914 Free PMC article. - EZH2, JMJD3, and UTX epigenetically regulate hepatic plasticity inducing retro-differentiation and proliferation of liver cells.
Pediconi N, Salerno D, Lupacchini L, Angrisani A, Peruzzi G, De Smaele E, Levrero M, Belloni L. Pediconi N, et al. Cell Death Dis. 2019 Jul 8;10(7):518. doi: 10.1038/s41419-019-1755-2. Cell Death Dis. 2019. PMID: 31285428 Free PMC article. - Role of H3K27 demethylases Jmjd3 and UTX in transcriptional regulation.
Hübner MR, Spector DL. Hübner MR, et al. Cold Spring Harb Symp Quant Biol. 2010;75:43-9. doi: 10.1101/sqb.2010.75.020. Epub 2011 Jan 5. Cold Spring Harb Symp Quant Biol. 2010. PMID: 21209387 - JMJD3 as an epigenetic regulator in development and disease.
Burchfield JS, Li Q, Wang HY, Wang RF. Burchfield JS, et al. Int J Biochem Cell Biol. 2015 Oct;67:148-57. doi: 10.1016/j.biocel.2015.07.006. Epub 2015 Jul 17. Int J Biochem Cell Biol. 2015. PMID: 26193001 Free PMC article. Review. - Lysine Demethylase KDM6A in Differentiation, Development, and Cancer.
Tran N, Broun A, Ge K. Tran N, et al. Mol Cell Biol. 2020 Sep 28;40(20):e00341-20. doi: 10.1128/MCB.00341-20. Print 2020 Sep 28. Mol Cell Biol. 2020. PMID: 32817139 Free PMC article. Review.
Cited by
- Critical role of histone demethylase Jmjd3 in the regulation of CD4+ T-cell differentiation.
Li Q, Zou J, Wang M, Ding X, Chepelev I, Zhou X, Zhao W, Wei G, Cui J, Zhao K, Wang HY, Wang RF. Li Q, et al. Nat Commun. 2014 Dec 22;5:5780. doi: 10.1038/ncomms6780. Nat Commun. 2014. PMID: 25531312 Free PMC article. - The histone demethylase jumonji coordinates cellular senescence including secretion of neural stem cell-attracting cytokines.
Perrigue PM, Silva ME, Warden CD, Feng NL, Reid MA, Mota DJ, Joseph LP, Tian YI, Glackin CA, Gutova M, Najbauer J, Aboody KS, Barish ME. Perrigue PM, et al. Mol Cancer Res. 2015 Apr;13(4):636-50. doi: 10.1158/1541-7786.MCR-13-0268. Epub 2015 Feb 4. Mol Cancer Res. 2015. PMID: 25652587 Free PMC article. - Kdm6b Regulates the Generation of Effector CD8+ T Cells by Inducing Chromatin Accessibility in Effector-Associated Genes.
Xu T, Schutte A, Jimenez L, Gonçalves ANA, Keller A, Pipkin ME, Nakaya HI, Pereira RM, Martinez GJ. Xu T, et al. J Immunol. 2021 May 1;206(9):2170-2183. doi: 10.4049/jimmunol.2001459. Epub 2021 Apr 16. J Immunol. 2021. PMID: 33863789 Free PMC article. - SMARCA4/Brg1 coordinates genetic and epigenetic networks underlying Shh-type medulloblastoma development.
Shi X, Wang Q, Gu J, Xuan Z, Wu JI. Shi X, et al. Oncogene. 2016 Nov 3;35(44):5746-5758. doi: 10.1038/onc.2016.108. Epub 2016 Apr 11. Oncogene. 2016. PMID: 27065321 - Dnmt3a silences hematopoietic stem cell self-renewal.
Trowbridge JJ, Orkin SH. Trowbridge JJ, et al. Nat Genet. 2011 Dec 27;44(1):13-4. doi: 10.1038/ng.1043. Nat Genet. 2011. PMID: 22200773
References
- Agger K, Christensen J, Cloos PAC, Helin K. The emerging functions of histone demethylases. Current Opinion in Genetics and Development. 2008;18:1–10. - PubMed
- Andreou AM, Pauws E, Jones MC, Singh MK, Bussen M, Doudney K, Moore GE, Kispert A, Brosens JJ, Stanier P. TBX22 missense mutations found in patients with X-linked cleft palate affect DNA binding, sumoylation, and transcriptional repression. American Journal of Human Genetics. 2007;81:700–712. - PMC - PubMed
- Beima KM, Miazgowicz MM, Lewis MD, Yan PS, Huang TH, Weinmann AS. T-bet binding to newly identified target gene promoters is cell type-independent but results in variable context-dependent functional effects. The Journal of Biological Chemistry. 2006;281:11992–12000. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- AI07272/AI/NIAID NIH HHS/United States
- R01 AI071272/AI/NIAID NIH HHS/United States
- 2T32 GM007270/GM/NIGMS NIH HHS/United States
- T32 GM066698/GM/NIGMS NIH HHS/United States
- T32 GM007270/GM/NIGMS NIH HHS/United States
- R01 AI061061-07/AI/NIAID NIH HHS/United States
- T32 AI007272/AI/NIAID NIH HHS/United States
- R01 AI061061/AI/NIAID NIH HHS/United States
- R01 AI071272-10/AI/NIAID NIH HHS/United States
- R56 AI061061/AI/NIAID NIH HHS/United States
- AI061061/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous