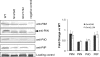The peptidoglycan-binding protein FimV promotes assembly of the Pseudomonas aeruginosa type IV pilus secretin - PubMed (original) (raw)
The peptidoglycan-binding protein FimV promotes assembly of the Pseudomonas aeruginosa type IV pilus secretin
Hania Wehbi et al. J Bacteriol. 2011 Jan.
Abstract
The Pseudomonas aeruginosa inner membrane protein FimV is among several proteins of unknown function required for type IV pilus-mediated twitching motility, arising from extension and retraction of pili from their site of assembly in the inner membrane. The pili transit the periplasm and peptidoglycan (PG) layer, ultimately exiting the cell through the PilQ secretin. Although fimV mutants are nonmotile, they are susceptible to killing by pilus-specific bacteriophage, a hallmark of retractable surface pili. Here we show that levels of recoverable surface pili were markedly decreased in fimV pilT retraction-deficient mutants compared with levels in the pilT control, demonstrating that FimV acts at the level of pilus assembly. Levels of inner membrane assembly subcomplex proteins PilM/N/O/P were decreased in fimV mutants, but supplementation of these components in trans did not restore pilus assembly or motility. Loss of FimV dramatically reduced the levels of the PilQ secretin multimer through which pili exit the cell, in part due to decreased levels of PilQ monomers, while PilF pilotin levels were unchanged. Expression of pilQ in trans in the wild type or fimV mutants increased total PilQ monomer levels but did not alter secretin multimer levels or motility. PG pulldown assays showed that the N terminus of FimV bound PG in a LysM motif-dependent manner, and a mutant with an in-frame chromosomal deletion of the LysM motif had reduced motility, secretin levels, and surface piliation. Together, our data show that FimV's role in pilus assembly is to promote secretin formation and that this function depends upon its PG-binding domain.
Figures
FIG. 1.
Schematic of FimV. The 919-residue FimV protein contains a peptidoglycan binding domain (LysM, blue), two Pro/Ala-rich regions (P/A, orange), a coiled-coil region (CC, green), a predicted transmembrane segment (TM, white), a highly acidic cytoplasmic domain (red) containing predicted TPR domains, and a C-terminal region of unknown function, also predicted to have TPR-like structure, that is highly conserved among FimV orthologues (purple). The positions of disruption in the fimV346 and fimV1194 mutants are indicated with arrowheads.
FIG. 2.
Analysis of sheared surface proteins using SDS-PAGE. (Top) Representative sheared surface protein preparation of wild-type (WT), pilA (nonpiliated, NP), pilT, and fimV mutant, and fimV pilT double mutant strains, visualized on a Coomassie-stained SDS-polyacrylamide gel (12.5%). The flagellin band was used as a loading control. The molecular size markers (in kilodaltons) are indicated on the left. (Bottom) Western blot analysis of the same samples probed using an anti-PilA antibody. The fimV mutants lack recoverable surface pili, and only a small number of pili can be recovered when retraction is blocked with a pilT mutation. The susceptibility of each strain to killing by PO4 bacteriophage is indicated at the bottom (+, phage susceptible; −, phage resistant).
FIG. 3.
Levels of PilM/N/O/P are decreased in fimV mutants. (Left) Representative Western blot of whole-cell lysates of the WT and the fimV mutants, probed using antibodies raised against PilM/N/O/P. The negative-control lane contains whole-cell lysate from the appropriate mutant (4). A cross-reactive band was included as a loading control. (Right) Results of densitometric analyses showing average fold changes in the levels of the indicated proteins in the fimV mutants compared to wild-type levels (set to 1). Results are the averages of 3 independent experiments; error bars indicate standard deviations.
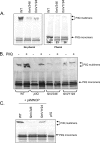
FIG. 4.
Levels of PilQ multimer are reduced in fimV mutants. (A) Representative Western blot of whole-cell lysates of the WT and the fimV mutants, probed using an antibody to PilQ (29). Samples on the right were treated with phenol to dissociate PilQ multimers. Numbers in the right panel indicate the amount (%) of monomer present in each strain following phenol dissociation as quantified by densitometry, with the wild-type amount set to 100%. (B) Whole-cell lysates of the wild type (WT) and the pilQ and fimV mutants, carrying the pBADGr vector control (−) or pBADGr-pilQ (+), probed with an antibody to PilQ. (C) Whole-cell lysates of the wild type (WT) and the fimV mutants, transformed with a low-copy-number plasmid expressing PilM/N/O/P, probed with an antibody to PilQ. The pilQ mutant was included as a negative control.
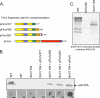
FIG. 5.
Complementation with FimV fragments. (A) Schematic of FimV fragments used to complement the fimV1194 mutant in panel B. The color coding is described in the legend for Fig. 1. (B) (Top) Western blot, probed with an anti-PilA antibody, of sheared surface proteins from the wild type (WT), the pilA mutant (nonpiliated [NP]), the fimV1194 mutant, and the fimV1194 mutant complemented with stable fragments of FimV. (Bottom) Representative twitching motility zones for each strain. Bar, 1 cm. (C) Western blot of whole-cell lysates of the wild type (WT) and the fimV mutants, probed with an antibody to the C-terminal fragment of FimV (residues 468 to 919). The fimV1194 mutant makes a stable, antibody-reactive fragment.
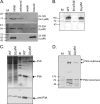
FIG. 6.
FimV's LysM domain is required for PG binding and secretin formation. (A) PG pulldown assay. Purified fragments (amino acid positions shown at right) of the N terminus of FimV were incubated with purified P. aeruginosa PG as described in Materials and Methods. Fragments containing the LysM motif (middle and bottom) bound PG, while a shorter fragment did not (top). Note that while the same amount of protein was used for all three fragments, the fragment from amino acids 23 to 390 has both N- and C-terminal His tags and therefore stains more intensely with the anti-His antibody used for detection. (B) Western blot of whole-cell lysates of the wild type (WT) and the fimV346 and ΔLysM mutants, probed with antibody to the C-terminal fragment of FimV (residues 468 to 919). (C) (Top) Coomassie blue-stained SDS-polyacrylamide gel of whole-cell lysates (left) and sheared surface proteins (right) from the wild type (WT) and the ΔLysM mutant. (Bottom) Western blot of the same samples probed with an antibody to PilA. (D) Western blot of the wild type (WT) and the ΔLysM mutant, probed with an anti-PilQ antibody. In each panel, M denotes the molecular size markers in kilodaltons.
References
- Ahn, K. S., U. Ha, J. Jia, D. Wu, and S. Jin. 2004. The truA gene of Pseudomonas aeruginosa is required for the expression of type III secretory genes. Microbiology 150**:**539-547. -PubMed
- Ayers, M., P. L. Howell, and L. L. Burrows. 2010. Architecture of the type II secretion and type IV pilus machineries. Future Microbiol. 5**:**1203-1218. -PubMed
- Ayers, M., et al. 2009. PilM/N/O/P proteins form an inner membrane complex that affects the stability of the Pseudomonas aeruginosa type IV pilus secretin. J. Mol. Biol. 394**:**128-142. -PubMed
- Bateman, A., and M. Bycroft. 2000. The structure of a LysM domain from E. coli membrane-bound lytic murein transglycosylase D (MltD). J. Mol. Biol. 299**:**1113-1119. -PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases