NCBI GEO: archive for functional genomics data sets--10 years on - PubMed (original) (raw)
. 2011 Jan;39(Database issue):D1005-10.
doi: 10.1093/nar/gkq1184. Epub 2010 Nov 21.
Dennis B Troup, Stephen E Wilhite, Pierre Ledoux, Carlos Evangelista, Irene F Kim, Maxim Tomashevsky, Kimberly A Marshall, Katherine H Phillippy, Patti M Sherman, Rolf N Muertter, Michelle Holko, Oluwabukunmi Ayanbule, Andrey Yefanov, Alexandra Soboleva
Affiliations
- PMID: 21097893
- PMCID: PMC3013736
- DOI: 10.1093/nar/gkq1184
NCBI GEO: archive for functional genomics data sets--10 years on
Tanya Barrett et al. Nucleic Acids Res. 2011 Jan.
Abstract
A decade ago, the Gene Expression Omnibus (GEO) database was established at the National Center for Biotechnology Information (NCBI). The original objective of GEO was to serve as a public repository for high-throughput gene expression data generated mostly by microarray technology. However, the research community quickly applied microarrays to non-gene-expression studies, including examination of genome copy number variation and genome-wide profiling of DNA-binding proteins. Because the GEO database was designed with a flexible structure, it was possible to quickly adapt the repository to store these data types. More recently, as the microarray community switches to next-generation sequencing technologies, GEO has again adapted to host these data sets. Today, GEO stores over 20,000 microarray- and sequence-based functional genomics studies, and continues to handle the majority of direct high-throughput data submissions from the research community. Multiple mechanisms are provided to help users effectively search, browse, download and visualize the data at the level of individual genes or entire studies. This paper describes recent database enhancements, including new search and data representation tools, as well as a brief review of how the community uses GEO data. GEO is freely accessible at http://www.ncbi.nlm.nih.gov/geo/.
Figures
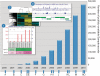
Figure 1.
A timeline of GEO database growth, development and events. The chart represents accumulative growth of publicly-available Sample records from 2000 to September 2010. A further 80 000 Samples are currently held private until published, making a total of about 550 000 Samples. The current rate of submission and processing is over 10 000 Samples per month. (A) First data uploaded to database. (B) MIAME proposal is published, outlining the minimum information that should be included when describing a microarray experiment (2). (C) Nature journals announce requirement for microarray data deposit to public databases (4). (D) Reviewer access mechanism enabled, allowing anonymous confidential review of pre-published data. (E and inset 1) GEO Profiles database released, enabling search and visualization of individual gene expression charts. (F and inset 2) Interactive pre-computed cluster heatmaps released, allowing users to view and select regions of interesting gene expression patterns. (G) Major database modifications released aimed at better support of MIAME elements. (H) GEO increases enforcement of provision of raw data. (I) Bioconductor GEOquery package published, allowing GEO data to be imported into R environment (22). (J) GEOarchive spreadsheet submission format released, enabling rapid batch deposit of data. (K) All GEO Series records re-classified according to technology and experiment type making it simple to locate studies of a specific type; types are listed in Table 1. (L) Improvements to DataSet Browser and accompanying analysis tools panel implemented. (M and inset 3) First release of next-generation sequence tracks on NCBI’s Sequence Viewer. These tracks were generated in support of the NIH Roadmap Epigenomics project,
http://www.ncbi.nlm.nih.gov/geo/roadmap/epigenomics/
. (N) Links generated to NCBI’s new Epigenomics resource (7) which applies advanced curation and genome browser tracks for hundreds of next-generation sequence Samples derived from GEO. (O) Advanced Search tool released, helping users construct complex queries.
Similar articles
- NCBI GEO: archive for high-throughput functional genomic data.
Barrett T, Troup DB, Wilhite SE, Ledoux P, Rudnev D, Evangelista C, Kim IF, Soboleva A, Tomashevsky M, Marshall KA, Phillippy KH, Sherman PM, Muertter RN, Edgar R. Barrett T, et al. Nucleic Acids Res. 2009 Jan;37(Database issue):D885-90. doi: 10.1093/nar/gkn764. Epub 2008 Oct 21. Nucleic Acids Res. 2009. PMID: 18940857 Free PMC article. - NCBI GEO: archive for functional genomics data sets--update.
Barrett T, Wilhite SE, Ledoux P, Evangelista C, Kim IF, Tomashevsky M, Marshall KA, Phillippy KH, Sherman PM, Holko M, Yefanov A, Lee H, Zhang N, Robertson CL, Serova N, Davis S, Soboleva A. Barrett T, et al. Nucleic Acids Res. 2013 Jan;41(Database issue):D991-5. doi: 10.1093/nar/gks1193. Epub 2012 Nov 27. Nucleic Acids Res. 2013. PMID: 23193258 Free PMC article. - NCBI GEO: mining tens of millions of expression profiles--database and tools update.
Barrett T, Troup DB, Wilhite SE, Ledoux P, Rudnev D, Evangelista C, Kim IF, Soboleva A, Tomashevsky M, Edgar R. Barrett T, et al. Nucleic Acids Res. 2007 Jan;35(Database issue):D760-5. doi: 10.1093/nar/gkl887. Epub 2006 Nov 11. Nucleic Acids Res. 2007. PMID: 17099226 Free PMC article. - Gene expression omnibus: microarray data storage, submission, retrieval, and analysis.
Barrett T, Edgar R. Barrett T, et al. Methods Enzymol. 2006;411:352-69. doi: 10.1016/S0076-6879(06)11019-8. Methods Enzymol. 2006. PMID: 16939800 Free PMC article. Review. - Comparative Genomics: Volumes 1 and 2.
Bergman NH, editor. Bergman NH, editor. Totowa (NJ): Humana Press; 2007. Totowa (NJ): Humana Press; 2007. PMID: 21250292 Free Books & Documents. Review.
Cited by
- An integrative approach to inferring gene regulatory module networks.
Baitaluk M, Kozhenkov S, Ponomarenko J. Baitaluk M, et al. PLoS One. 2012;7(12):e52836. doi: 10.1371/journal.pone.0052836. Epub 2012 Dec 20. PLoS One. 2012. PMID: 23285197 Free PMC article. - EPD and EPDnew, high-quality promoter resources in the next-generation sequencing era.
Dreos R, Ambrosini G, Cavin Périer R, Bucher P. Dreos R, et al. Nucleic Acids Res. 2013 Jan;41(Database issue):D157-64. doi: 10.1093/nar/gks1233. Epub 2012 Nov 27. Nucleic Acids Res. 2013. PMID: 23193273 Free PMC article. - BioGPS and GXD: mouse gene expression data-the benefits and challenges of data integration.
Ringwald M, Wu C, Su AI. Ringwald M, et al. Mamm Genome. 2012 Oct;23(9-10):550-8. doi: 10.1007/s00335-012-9408-0. Epub 2012 Jul 31. Mamm Genome. 2012. PMID: 22847375 Free PMC article. - Network pharmacology strategies toward multi-target anticancer therapies: from computational models to experimental design principles.
Tang J, Aittokallio T. Tang J, et al. Curr Pharm Des. 2014;20(1):23-36. doi: 10.2174/13816128113199990470. Curr Pharm Des. 2014. PMID: 23530504 Free PMC article. Review. - YM500: a small RNA sequencing (smRNA-seq) database for microRNA research.
Cheng WC, Chung IF, Huang TS, Chang ST, Sun HJ, Tsai CF, Liang ML, Wong TT, Wang HW. Cheng WC, et al. Nucleic Acids Res. 2013 Jan;41(Database issue):D285-94. doi: 10.1093/nar/gks1238. Epub 2012 Nov 29. Nucleic Acids Res. 2013. PMID: 23203880 Free PMC article.
References
- Brazma A, Hingamp P, Quackenbush J, Sherlock G, Spellman P, Stoeckert C, Aach J, Ansorge W, Ball CA, Causton HC, et al. Minimum information about a microarray experiment (MIAME)-toward standards for microarray data. Nat. Genet. 2001;29:365–371. - PubMed
- Microarray standards at last. Nature. 2002;419:323. Available at http://www.nature.com/nature/journal/v419/n6905/full/419323a.html. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources