Sox10 directs neural stem cells toward the oligodendrocyte lineage by decreasing Suppressor of Fused expression - PubMed (original) (raw)
Sox10 directs neural stem cells toward the oligodendrocyte lineage by decreasing Suppressor of Fused expression
Christine D Pozniak et al. Proc Natl Acad Sci U S A. 2010.
Abstract
Oligodendrocyte precursor cells (OPCs) are lineage-restricted progenitors generally limited in vivo to producing oligodendrocytes. Mechanisms controlling genesis of OPCs are of interest because of their importance in myelin development and their potential for regenerative therapies in multiple sclerosis and dysmyelinating syndromes. We show here that the SoxE transcription factors (comprising Sox8, 9, and 10) induce multipotent neural precursor cells (NPCs) from the early postnatal subventricular zone (SVZ) to become OPCs in an autonomous manner. We performed a chromatin immunoprecipitation-based bioinformatic screen and identified Suppressor of Fused (Sufu) as a direct target of repression by Sox10. In vitro, overexpression of Sufu blocked OPC production, whereas RNAi-mediated inhibition augmented OPC production. Furthermore, mice heterozygous for Sufu have increased numbers of OPCs in the telencephalon during development. We conclude that Sox10 acts to restrict the potential of NPCs toward the oligodendrocyte lineage in part by regulating the expression of Sufu.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
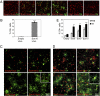
Fig. 1.
Overexpression of SoxE factors but not Olig genes is sufficient to drive differentiation toward the oligodendrocyte lineage. (A) Empty virus infected cells do not express O4 in EGF/FGF, whereas Sox8, Sox9, or Sox10 virus-infected cells express O4 at significant levels. In contrast, Olig1 or Olig2 expression does not lead to increased O4 staining in these culture conditions. (B) The efficiency of O4+ cell production after Sox10 expression under nondifferentiating conditions (in EGF/FGF containing medium). Results pooled from three separate experiments. Error bars are ± SEM, ***P < 0.005. (_C_) Cells infected with either Empty or SoxE-expressing retroviruses (red) cultured in 2% FBS for 48 h and stained with O4 antibody (green). _Inset_ shows Sox10-infected cells also stain with CNPase, a marker of terminally differentiating oligodendrocytes. (_D_) Cells infected with either Empty or SoxE-expressing retroviruses (red) cultured in thyroid hormones for 48 h and stained with O4 antibody (green). _Inset_ shows Sox10-infected cells also stain with CNPase, a marker of terminally differentiating oligodendrocytes. (_E_) The percentage of O4+ cells infected with Sox8, 9, or 10 cultured in serum or thyroid hormones for 48 h. Results are pooled from >3 separate experiments; error bars are ±SEM; *P < 0.05; **P < 0.02; ***P < 0.005.
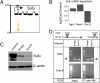
Fig. 2.
Suppressor of Fused (Sufu) is a direct target of Sox10. (A) Schematic of the Sufu locus depicting the Sox10-binding site identified in the first intron of the Sufu gene (
SI Methods
). (B) Quantitative mRNA profiling using whole mouse transcriptome Affymetrix chips. Each value is shown as a ratio of the experimental value over Empty virus-infected controls. Shown are the relative expression of Sufu mRNA 48 h after infection with Sox10 virus or 24 h after infection with Mash1 or Ngn2 retroviruses that drive neurogenic differentiation in EGF/FGF containing medium. Note Mash1 and Ngn2 drive modest increases in Sufu expression, whereas Sox10 leads to a twofold decrease in expression of Sufu. Normalized intensity values of triplicate chip hybridization experiments are presented. Each sample was prepared from pools of mRNA derived from less than three separate experiments. (C) Sox10 expression leads to decreased Sufu protein levels in NPCs 24 h after plating. Note the slightly higher and broader band in the positive control lane was obtained from extracts of HEK-293T cells expressing both human and mouse HA-tagged Sufu. In contrast, NPCs only express endogenous untagged mouse Sufu. (D) Targeted ChIP for Sox10 bound to the Sufu gene at the first intronic site. Positions of the PCR primers are shown in relationship to the Sufu and Trim8 genes. The PCR yields the expected products for both Sufu and Trim8 after infection with either Empty or Sox10 virus (Input), but only Sox10-infected cells yielded a specific band for Sufu after ChIP (IP: anti-HA). Empty virus-infected cells showed no signal, and Sox10 showed no binding to the closely linked Trim8 gene. The arrows indicate the size of the expected Sufu product.
Fig. 3.
Sufu inhibits Sox10-driven OPC differentiation and endogenous oligodendrocyte differentiation. (A) shRNA-mediated knock down of mouse HA-Sufu protein (solid arrow, top bands) in GP2-293 cells. Note that the endogenous human Sufu protein is unaffected (open arrow, bottom bands). (B) Percentage of O4+ cells coinfected with Sox10 and Luciferase shRNA, Sufu, or Sufu siRNA. Transduced NPCs were cultured in EGF/FGF on laminin-coated plates for 72 h. Error bars are ± SEM, ***P < 0.005, n = 3. (C) Percentage of O4+ cells coinfected with Sox10 virus, and either Empty or Sufu virus then induced to differentiate by removing EGF/FGF and addition of serum. Error bars are ±SEM, **P < 0.01, n = 3. (D) Percentage of O4+ cells infected with Empty or Sufu virus and cultured in serum on laminin-coated plates for 72 h. Error bars are ± SEM, ***P < 0.01, n = 3.
Fig. 4.
Decreased Sufu expression significantly increases the number of oligodendrocyte precursors in vivo. (A) Western blot analysis of equal amounts of protein derived from brain lysates of wild-type (Sufu+/+) mice or mice lacking one copy of Sufu (Sufu+/−). (B) Olig2 staining in brain sections of Sufu+/+ or Sufu+/− E15.5 brains show an overall increase in Olig2-positive cells in Sufu+/− brains. Higher magnifications in Lower. (Scale bar: 100 μm.) (C) The number of PDGFRα-positive cells in the forebrain is significantly increased in mice with decreased Sufu expression (Sufu+/−) at E15.5 (Error bars are ±SD, ***P < 0.005, n = 3). (D) The increased number of PDGFRα-positive cells persists in the Sufu+/− P2 forebrain. Error bars are ± SD, **P < 0.05, n = 3.
Similar articles
- High purity of human oligodendrocyte progenitor cells obtained from neural stem cells: suitable for clinical application.
Wang C, Luan Z, Yang Y, Wang Z, Wang Q, Lu Y, Du Q. Wang C, et al. J Neurosci Methods. 2015 Jan 30;240:61-6. doi: 10.1016/j.jneumeth.2014.10.017. Epub 2014 Nov 8. J Neurosci Methods. 2015. PMID: 25445251 - Sox10 is necessary for oligodendrocyte survival following axon wrapping.
Takada N, Kucenas S, Appel B. Takada N, et al. Glia. 2010 Jun;58(8):996-1006. doi: 10.1002/glia.20981. Glia. 2010. PMID: 20229602 Free PMC article. - Gsx transcription factors control neuronal versus glial specification in ventricular zone progenitors of the mouse lateral ganglionic eminence.
Chapman H, Riesenberg A, Ehrman LA, Kohli V, Nardini D, Nakafuku M, Campbell K, Waclaw RR. Chapman H, et al. Dev Biol. 2018 Oct 1;442(1):115-126. doi: 10.1016/j.ydbio.2018.07.005. Epub 2018 Jul 7. Dev Biol. 2018. PMID: 29990475 Free PMC article. - Intrinsic and extrinsic control of oligodendrocyte development.
Zuchero JB, Barres BA. Zuchero JB, et al. Curr Opin Neurobiol. 2013 Dec;23(6):914-20. doi: 10.1016/j.conb.2013.06.005. Epub 2013 Jul 3. Curr Opin Neurobiol. 2013. PMID: 23831087 Free PMC article. Review. - Oligodendrogenesis from neural stem cells: perspectives for remyelinating strategies.
Grade S, Bernardino L, Malva JO. Grade S, et al. Int J Dev Neurosci. 2013 Nov;31(7):692-700. doi: 10.1016/j.ijdevneu.2013.01.004. Epub 2013 Jan 20. Int J Dev Neurosci. 2013. PMID: 23340483 Review.
Cited by
- Rapid generation of OPC-like cells from human pluripotent stem cells for treating spinal cord injury.
Kim DS, Jung SJ, Lee JS, Lim BY, Kim HA, Yoo JE, Kim DW, Leem JW. Kim DS, et al. Exp Mol Med. 2017 Jul 28;49(7):e361. doi: 10.1038/emm.2017.106. Exp Mol Med. 2017. PMID: 28751784 Free PMC article. - A novel method for culturing enteric neurons generates neurospheres containing functional myenteric neuronal subtypes.
Mandal A, Moneme C, Tewari BP, Goldstein AM, Sontheimer H, Cheng L, Moore SR, Levin D. Mandal A, et al. J Neurosci Methods. 2024 Jul;407:110144. doi: 10.1016/j.jneumeth.2024.110144. Epub 2024 Apr 25. J Neurosci Methods. 2024. PMID: 38670535 Free PMC article. - Short-range Fgf signalling patterns hindbrain progenitors to induce the neurogenesis-to-oligodendrogenesis switch.
Yeung TJ, Wilkinson DG. Yeung TJ, et al. Development. 2024 Dec 15;151(24):dev204256. doi: 10.1242/dev.204256. Epub 2024 Dec 13. Development. 2024. PMID: 39575980 Free PMC article. - Human iPSC-Derived Neural Models for Studying Alzheimer's Disease: from Neural Stem Cells to Cerebral Organoids.
Barak M, Fedorova V, Pospisilova V, Raska J, Vochyanova S, Sedmik J, Hribkova H, Klimova H, Vanova T, Bohaciakova D. Barak M, et al. Stem Cell Rev Rep. 2022 Feb;18(2):792-820. doi: 10.1007/s12015-021-10254-3. Epub 2022 Feb 2. Stem Cell Rev Rep. 2022. PMID: 35107767 Free PMC article. Review. - Sonic Hedgehog Signaling Rises to the Surface: Emerging Roles in Neocortical Development.
Yabut OR, Pleasure SJ. Yabut OR, et al. Brain Plast. 2018 Aug 10;3(2):119-128. doi: 10.3233/BPL-180064. Brain Plast. 2018. PMID: 30151337 Free PMC article. Review.
References
- Wilson SW, Rubenstein JL. Induction and dorsoventral patterning of the telencephalon. Neuron. 2000;28:641–651. - PubMed
- Briscoe J, Ericson J. Specification of neuronal fates in the ventral neural tube. Curr Opin Neurobiol. 2001;11:43–49. - PubMed
- Tekki-Kessaris N, et al. Hedgehog-dependent oligodendrocyte lineage specification in the telencephalon. Development. 2001;128:2545–2554. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 NS039278/NS/NINDS NIH HHS/United States
- R21NS055783/NS/NINDS NIH HHS/United States
- P50 CA97257/CA/NCI NIH HHS/United States
- P50 CA097257/CA/NCI NIH HHS/United States
- R21 NS055783/NS/NINDS NIH HHS/United States
- K02 MH074958/MH/NIMH NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous