Combining ATR suppression with oncogenic Ras synergistically increases genomic instability, causing synthetic lethality or tumorigenesis in a dosage-dependent manner - PubMed (original) (raw)
Combining ATR suppression with oncogenic Ras synergistically increases genomic instability, causing synthetic lethality or tumorigenesis in a dosage-dependent manner
Oren Gilad et al. Cancer Res. 2010.
Abstract
Previous studies indicate that oncogenic stress activates the ATR-Chk1 pathway. Here, we show that ATR-Chk1 pathway engagement is essential for limiting genomic instability following oncogenic Ras transformation. ATR pathway inhibition in combination with oncogenic Ras expression synergistically increased genomic instability, as quantified by chromatid breaks, sister chromatid exchanges, and H2AX phosphorylation. This level of instability was significantly greater than that observed following ATR suppression in untransformed control cells. In addition, consistent with a deficiency in long-term genome maintenance, hypomorphic ATR pathway reduction to 16% of normal levels was synthetic lethal with oncogenic Ras expression in cultured cells. Notably, elevated genomic instability and synthetic lethality following suppression of ATR were not due to accelerated cycling rates in Ras-transformed cells, indicating that these synergistic effects were generated on a per-cell-cycle basis. In contrast to the synthetic lethal effects of hypomorphic ATR suppression, subtle reduction of ATR expression (haploinsufficiency) in combination with endogenous levels of K-ras(G12D) expression elevated the incidence of lung adenocarcinoma, spindle cell sarcoma, and thymic lymphoma in p53 heterozygous mice. K-ras(G12D)-induced tumorigenesis in ATR(+/-)p53(+/-) mice was associated with intrachromosomal deletions and loss of wild-type p53. These findings indicate that synergistic increases in genomic instability following ATR reduction in oncogenic Ras-transformed cells can produce 2 distinct biological outcomes: synthetic lethality upon significant suppression of ATR expression and tumor promotion in the context of ATR haploinsufficiency. These results highlight the importance of the ATR pathway both as a barrier to malignant progression and as a potential target for cancer treatment.
Conflict of interest statement
Disclosure of Potential Conflicts of Interest
None
Figures
Figure 1
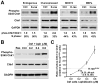
Expression of oncogenic Ras mutants activates the ATR-Chk1 pathway. A, Western blot detection of Chk1-S345 phosphorylation in normal and oncogenic Ras-transformed cells. Control and K-rasG12D- or H-rasG12V-expressing murine embryonic fibroblast lines grown in defined media (Materials and Methods) were detected for Chk1-S345 phosphorylation by western blot. The increase in phospho-Chk1 over total Chk1 abundance was calculated on each blot relative to the levels observed in untransformed controls. GAPDH was additionally detected and quantified as a loading control. Flow cytometric quantification of S phase representation was performed in parallel by 45-minute BrdU incorporation and propidium iodide (PI) staining. B, Western blot detection of Chk1 phosphorylation in H-rasG12V-transformed NIH3T3 cells in comparison to control cells treated for one hour with increasing concentrations of aphidicolin. C, Quantification of aphidicolin-stimulated Chk1 phosphorylation relative to average H-rasG12V-stimulated phosphorylation. Western blots and quantification of Chk1 phosphorylation are representative of 3–4 independent experiments. Standard error (SE) bars are shown.
Figure 2
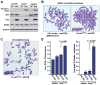
Oncogenic Ras expression in combination with ATR-Chk1 pathway suppression leads to increased genomic instability. A, Increased H2AX phosphorylation upon ATR-Chk1 pathway suppression in combination with H-rasG12V-transformation. ATR-Chk1 pathway was inhibited in H-rasG12V-transformed or untransformed control cells (pBabe-puro transduced) via shRNA-mediated reduction of ATR expression (21) or three-hour treatment with the Chk1 kinase inhibitor Gö6976. Lysates were detected for H2AX phosphorylation by western blot; Ras and ATR levels were also detected with GAPDH and MCM3 as loading controls, respectively. Expression of shATR reduced ATR protein levels by 94% (untransformed control cells) and 81% (H-rasG12V-transformed cells) in the experiment shown. These values were within the typical range of ATR reduction [86.9% ± 5.6 (S.E.) for untransformed controls, and 81.7% ± 1.8 (S.E.) for H-rasG12V-transformed] and were sufficient to limit Chk1 S345 phosphorylation in response to low-dose aphidicolin (Supplemental Fig. 1). B and C, Representative SCE and chromatid break detection in H-rasG12V-transformed cells following shRNA-mediated ATR reduction. Mitotic spreads for SCE and chromatid break detection were collected 48 hours after infection with lentiviruses that expressed the indicated shRNAs. D, Quantification of average SCEs and chromatid gaps and breaks following ATR suppression in H-rasG12V-transformed and control cells. Data shown are derived from 3–5 independent experiments. For section D, standard error bars are shown and P values were calculated by the Student’s t test.
Figure 3
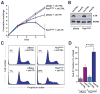
ATR-Chk1 pathway hypomorphic suppression is synthetic lethal with oncogenic Ras-transformation. A, Proliferation of H-rasG12V-transformed and control cells following ATR suppression. shRNA-mediated reduction of ATR was performed as described in Fig. 2. Cells were maintained at subconfluence by replating every two days, and cumulative population doublings were quantified over a total of 8 days. B, Western blot detection of ATR in lysates from control and H-rasG12V-transformed cells following shRNA-mediated ATR suppression (Day 2). C, Cell cycle profiles (PI staining) of control and H-rasG12V-transformed cells following ATR suppression. Sub-G1 populations are indicated (brackets). D, Quantification of average sub-G1 population frequency in H-rasG12V-transformed and control cells with or without ATR suppression. Data shown are derived from 3–6 independent experiments. For sections A and D, standard error bars are shown and P values were calculated by the Student’s t test.
Figure 4
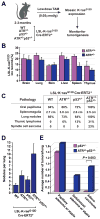
ATR haploinsufficiency promotes K-rasG12D-induced tumorigenesis. A, Schematic for ubiquitous mosaic activation of the lox-stop-lox (LSL) knock-in allele of K-rasG12D in ATR and p53 haploinsufficient mice. Recombination of the LSL-K-rasG12D allele was achieved through low-dose tamoxifen (TAM) activation of ubiquitously expressed Cre-ERT2 (28). B, Quantification of mosaic lox recombination of the LSL-K-rasG12D allele in various tissues. Real-time quantitative PCR (qPCR) analysis of genomic DNA was performed on tissues isolated from LSL-K-rasG12D/+Cre-ERT2+ mice (ATR+/+, n=3 and ATR+/−, n=6) five days after low-dose TAM treatment. Standard errors are indicated (bars). C, Tumorigenesis in ATR+/−p53+/− and control mice following Cre-mediated mosaic expression of K-rasG12D. The LSL-K-rasG12D/+Cre-ERT2+ mice analyzed were ATR+/+ (n=18), ATR+/− (n=17), p53+/− (n=20), and ATR+/−p53+/− (n=14). Tumors were identified upon necropsy or following IACUC-determined euthanasia endpoints, and the percentages of affected mice are shown. D, Average number of grossly-apparent lung nodules following mosaic activation of the LSL-K-rasG12D allele in ATR and p53 haploinsufficient mice. Nodules were quantified on fixed lungs isolated from low dose TAM-treated LSL-K-rasG12D/+Cre-ERT2+ mice with the indicated heterozygous deletions in ATR and p53 (n > 13 mice per genotype). E, ATR haploinsufficiency promotes K-rasG12D-induced lung adenocarcinoma in p53 heterozygous mice. Quantification of adenoma subtypes and adenocarcinoma was performed on lung tissue isolated from low dose TAM-treated LSL-K-rasG12D/+Cre-ERT2+ mice with the additional genotypes indicated. Blinded analysis of hematoxylin-eosin stained serial sectioning (1 per 50 microns, 24-39 sections in total were analyzed per lung) was performed, and adenoma subtypes and adenocarcinoma were classified as described (29). Standard errors (bars) were calculated from three or more independent PCR reactions (section B) or from nodules per lung (n > 13 mice, section D). P value (section E) was calculated by Fisher’s exact test.
Figure 5
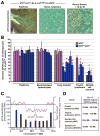
ATR haploinsufficiency accelerates p53 LOH in K-rasG12D-induced tumors. A, Detection of p53 by immunohistochemistry (IHC) in representative skin papilloma (n=2) and thymic lymphoma (n=4) isolated from low-dose TAM-treated ATR+/−p53+/−LSL-K-rasG12D/+Cre-ERT2+ mice. Normal thymus isolated from mice eight hours after exposure to 10 Gy ionizing radiation (IR) is shown as a positive control for p53 detection. Sections were stained in parallel using equivalent conditions. Nuclear p53 protein was not detected in spindle cell sarcomas isolated from ATR+/−p53+/− mice (n=3, data not shown). B, Detection of genomic loss of the wild-type p53 allele in tumors isolated from TAM-treated p53+/−LSL-K-rasG12D/+Cre-ERT2+ and ATR+/−p53+/−LSL-K-rasG12D/+Cre-ERT2+ mice. qPCR analysis of genomic DNA from tumors was performed as described in Materials and Methods. Wild-type p53 allele representation is shown relative to homozygous wild-type levels (p53+/+). Standard errors (bars) were calculated from technical replicates. Tumor samples in which the wild-type p53 allele frequency was significantly reduced are indicated (red asterisks). Enrichment of K-rasG12D expressing cells in tumor isolates was determined by qPCR quantification of LSL-K-rasG12D lox recombination (Supplemental Fig. 5). C, Detection of intrachromosomal deletion of p53 by array CGH and qPCR analysis. Representative qPCR regional quantification (blue bars) and array-CGH analysis of chromosome 11 (magenta line) on thymic lymphoma DNA isolated from a TAM-treated ATR+/−p53+/−LSL-K-rasG12D/+Cre-ERT2+ mouse. The location of the p53 gene is indicated. D, Deleted regions predicted by qPCR analysis of chromosome 11 in representative thymic lymphomas and spindle cell sarcomas from ATR+/−p53+/−LSL-K-rasG12D/+Cre-ERT2+ mice. Real-time primer sets (Supplemental Table 1) detecting chromosome 11 regions indicated in C were utilized in qPCR quantifications of tumor DNA and compared to pre-TAM treatment tail DNA. Regions that were selectively reduced in tumor DNA are shown.
Similar articles
- Oncogenic stress sensitizes murine cancers to hypomorphic suppression of ATR.
Schoppy DW, Ragland RL, Gilad O, Shastri N, Peters AA, Murga M, Fernandez-Capetillo O, Diehl JA, Brown EJ. Schoppy DW, et al. J Clin Invest. 2012 Jan;122(1):241-52. doi: 10.1172/JCI58928. Epub 2011 Dec 1. J Clin Invest. 2012. PMID: 22133876 Free PMC article. - ATR-Chk2 signaling in p53 activation and DNA damage response during cisplatin-induced apoptosis.
Pabla N, Huang S, Mi QS, Daniel R, Dong Z. Pabla N, et al. J Biol Chem. 2008 Mar 7;283(10):6572-83. doi: 10.1074/jbc.M707568200. Epub 2007 Dec 27. J Biol Chem. 2008. PMID: 18162465 - ATR and H2AX cooperate in maintaining genome stability under replication stress.
Chanoux RA, Yin B, Urtishak KA, Asare A, Bassing CH, Brown EJ. Chanoux RA, et al. J Biol Chem. 2009 Feb 27;284(9):5994-6003. doi: 10.1074/jbc.M806739200. Epub 2008 Dec 2. J Biol Chem. 2009. PMID: 19049966 Free PMC article. - The ATM-Chk2 and ATR-Chk1 pathways in DNA damage signaling and cancer.
Smith J, Tho LM, Xu N, Gillespie DA. Smith J, et al. Adv Cancer Res. 2010;108:73-112. doi: 10.1016/B978-0-12-380888-2.00003-0. Adv Cancer Res. 2010. PMID: 21034966 Review. - p53: guardian of the genome and policeman of the oncogenes.
Efeyan A, Serrano M. Efeyan A, et al. Cell Cycle. 2007 May 2;6(9):1006-10. doi: 10.4161/cc.6.9.4211. Epub 2007 May 28. Cell Cycle. 2007. PMID: 17457049 Review.
Cited by
- Targeting radioresistant breast cancer cells by single agent CHK1 inhibitor via enhancing replication stress.
Zhang Y, Lai J, Du Z, Gao J, Yang S, Gorityala S, Xiong X, Deng O, Ma Z, Yan C, Susana G, Xu Y, Zhang J. Zhang Y, et al. Oncotarget. 2016 Jun 7;7(23):34688-702. doi: 10.18632/oncotarget.9156. Oncotarget. 2016. PMID: 27167194 Free PMC article. - Perspectives on the combination of radiotherapy and targeted therapy with DNA repair inhibitors in the treatment of pancreatic cancer.
Yang SH, Kuo TC, Wu H, Guo JC, Hsu C, Hsu CH, Tien YW, Yeh KH, Cheng AL, Kuo SH. Yang SH, et al. World J Gastroenterol. 2016 Aug 28;22(32):7275-88. doi: 10.3748/wjg.v22.i32.7275. World J Gastroenterol. 2016. PMID: 27621574 Free PMC article. Review. - Inhibition of MEK and ATR is effective in a B-cell acute lymphoblastic leukemia model driven by Mll-Af4 and activated Ras.
Chu SH, Song EJ, Chabon JR, Minehart J, Matovina CN, Makofske JL, Frank ES, Ross K, Koche RP, Feng Z, Xu H, Krivtsov A, Nussenzweig A, Armstrong SA. Chu SH, et al. Blood Adv. 2018 Oct 9;2(19):2478-2490. doi: 10.1182/bloodadvances.2018021592. Blood Adv. 2018. PMID: 30266823 Free PMC article. - The Influence of Oncogenic RAS on Chemotherapy and Radiotherapy Resistance Through DNA Repair Pathways.
Cáceres-Gutiérrez RE, Alfaro-Mora Y, Andonegui MA, Díaz-Chávez J, Herrera LA. Cáceres-Gutiérrez RE, et al. Front Cell Dev Biol. 2022 Mar 11;10:751367. doi: 10.3389/fcell.2022.751367. eCollection 2022. Front Cell Dev Biol. 2022. PMID: 35359456 Free PMC article. Review. - Targeting ATR and Chk1 kinases for cancer treatment: a new model for new (and old) drugs.
Toledo LI, Murga M, Fernandez-Capetillo O. Toledo LI, et al. Mol Oncol. 2011 Aug;5(4):368-73. doi: 10.1016/j.molonc.2011.07.002. Epub 2011 Jul 28. Mol Oncol. 2011. PMID: 21820372 Free PMC article. Review.
References
- Gorgoulis VG, Vassiliou LV, Karakaidos P, et al. Activation of the DNA damage checkpoint and genomic instability in human precancerous lesions. Nature. 2005;434:907–913. - PubMed
- Bartkova J, Horejsi Z, Koed K, et al. DNA damage response as a candidate anti-cancer barrier in early human tumorigenesis. Nature. 2005;434:864–870. - PubMed
- Di Micco R, Fumagalli M, Cicalese A, et al. Oncogene-induced senescence is a DNA damage response triggered by DNA hyper-replication. Nature. 2006;444:638–642. - PubMed
- Bartkova J, Rezaei N, Liontos M, et al. Oncogene-induced senescence is part of the tumorigenesis barrier imposed by DNA damage checkpoints. Nature. 2006;444:633–637. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 AG027376-04S1/AG/NIA NIH HHS/United States
- R25CA101871/CA/NCI NIH HHS/United States
- R01 AG027376-04/AG/NIA NIH HHS/United States
- R01AG027376/AG/NIA NIH HHS/United States
- R01 AG027376/AG/NIA NIH HHS/United States
- R25 CA101871/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous