ABL fusion oncogene transformation and inhibitor sensitivity are mediated by the cellular regulator RIN1 - PubMed (original) (raw)
. 2011 Feb;25(2):290-300.
doi: 10.1038/leu.2010.268. Epub 2010 Nov 19.
Affiliations
- PMID: 21102429
- PMCID: PMC3049868
- DOI: 10.1038/leu.2010.268
Free PMC article
ABL fusion oncogene transformation and inhibitor sensitivity are mediated by the cellular regulator RIN1
M Thai et al. Leukemia. 2011 Feb.
Free PMC article
Abstract
ABL gene translocations create constitutively active tyrosine kinases that are causative in chronic myeloid leukemia, acute lymphocytic leukemia and other hematopoietic malignancies. Consistent retention of ABL SH3/SH2 autoinhibitory domains, however, suggests that these leukemogenic tyrosine kinase fusion proteins remain subject to regulation. We resolve this paradox, demonstrating that BCR-ABL1 kinase activity is regulated by RIN1, an ABL SH3/SH2 binding protein. BCR-ABL1 activity was increased by RIN1 overexpression and decreased by RIN1 silencing. Moreover, Rin1(-/-) bone marrow cells were not transformed by BCR-ABL1, ETV6-ABL1 or BCR-ABL1(T315I), a patient-derived drug-resistant mutant, as judged by growth factor independence. Rescue by ectopic RIN1 verified a cell autonomous mechanism of collaboration with BCR-ABL1 during transformation. Sensitivity to the ABL kinase inhibitor imatinib was increased by RIN1 silencing, consistent with RIN1 stabilization of an activated BCR-ABL1 conformation having reduced drug affinity. The dependence on activation by RIN1 to unleash full catalytic and cell transformation potential reveals a previously unknown vulnerability that could be exploited for treatment of leukemic cases driven by ABL translocations. The findings suggest that RIN1 targeting could be efficacious for imatinib-resistant disease and might complement ABL kinase inhibitors in first-line therapy.
Figures
Figure 1
BCR-ABL1 kinase activity in K562 cells is increased by RIN1. (a) Linear representation of ABL1 (top), translocation-derived human ABL1 and ABL2 fusion oncoproteins (middle six entries) and murine retroviral v-Abl (bottom). Src homology domains (SH2 and SH3) and tyrosine kinase (TK) domains are indicated. BCR-ABL1a represents the p190 isoform associated predominantly with ALL; BCR-ABL1b represents the p210 isoform associated predominantly with CML. (b) K562 cells transduced with vector (V) or RIN1 expression lentivirus (R) were treated with imatinib for 30 min at the indicated concentration and analyzed by immunoblot with anti-phosphotyrosine (MW markers in kDa at left). The ∼95 kDa band that intensifies in the R sample is most likely RIN1. β-tubulin (TUBB) immunoblot was used for normalization. (c) Levels of BCR-ABL1 (210 kDa) expression were evaluated by anti-ABL1 immunoblot of control (vector) and RIN1 overexpression (RIN1) K562 cells (ABL1 migrates below this region). RIN1 immunoblot showed ∼8- to 10-fold overexpression above endogenous levels. TUBB immunoblot was used to normalize extracts. (d) Tyrosine-phosphorylated endogenous CRKL (pY-CRKL) evaluated by immunoprecipitation with anti-CRKL and immunoblot with anti-phosphotyrosine (top) or anti-CRKL (bottom).
Figure 2
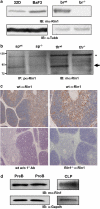
Endogenous mouse Rin1 expression. (a) Left: Immunoblot (IB) of Rin1 in 32D and BaF3 cells. Right: IB of wild-type (wt) and Rin1−/− (−/−) brain (br) tissue confirming antibody specificity. β-tubulin (Tubb) control below each blot. (b) IB of immunoprecipitated (IP) material from spleen (sp) and thymus (th) tissue of wt and Rin1−/− mice. Arrow indicates Rin1. Asterisk marks background band. mα, monoclonal antibody; pα, polyclonal antibody. (c) Immunohistochemical stain of Rin1 in mouse thymus. Top two panels show wild-type thymus probed with anti-Rin1 and counter stained with hematoxylin (left, × 4; right, × 10). Bottom left panel shows wild-type control without anti-Rin1. Bottom right panel shows Rin1−/− thymus control. (d) IB of Rin1 in PreB, ProB and common lymphoid progenitor (CLP) cells. Gapdh control below.
Figure 3
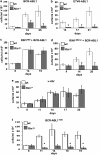
Rin1 is required for transformation of primary bone marrow (BM) cells to growth factor independence. (a) BM cells from wild-type or Rin1−/− mice were infected with a BCR-ABL1 (p210) retrovirus, cultured without growth factors and counted at indicated times. (b) BM cell transformation as in ‘a', except using ETV6-ABL1 (a.k.a. TEL-ABL) retrovirus. (c) BM cell transformation as in ‘a', except using virus expressing both BCR-ABL1 and RIN1ABD. (d) BM cell transformation as in ‘c', except using a mutant (RIN1ABDmut) that does not bind ABL1. (e) BM cell transformation as in ‘a', except using v-Abl retrovirus. (f) BM cell transformation as in ‘a', except using the multidrug-resistant mutant BCR-ABL1T315I. All results are for duplicate samples counted in triplicate. Bars show standard deviation; * indicates P<0.05 between wt and Rin1−/−. Panel f shows 14-day sample _P_=0.06 between wt and Rin1−/−.
Figure 4
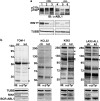
Analysis of human leukemia cells. (a) Panel of CML and ALL cell lines immunoblotted with anti-ABL1 (top), anti-RIN1 (middle) or anti-TUBB (bottom). CML cells (1=KCL22; 2=JURL-MK1; 3=K562) and a B lymphoid CML blast crisis cell line (4=BV173) express the p210 form of BCR-ABL, whereas ALL-derived cells (5=TOM-1; 6=SUP-B15) express the p190 form (arrowheads). Full-length RIN1 is marked with an arrow (faster migrating bands may be alternately spliced isoforms). (b) TOM-1, KCL22, K562 and primary pre-B-ALL cells infected with control (ctr) or RIN1-directed (kd) shRNA were analyzed by immunoblot with anti-phosphotyrosine. MW (kDa) markers are at left, arrowheads mark bands most clearly reduced by RIN1 silencing. Lower panels show immunoblots for TUBB, RIN1 and BCR-ABL1.
Figure 5
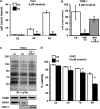
RIN1 silencing sensitizes ALL cells to imatinib. (a) Control (ctr) and RIN1-silenced (kd) TOM-1 cells (1 × 104/ml) were cultured in 8 μ imatinib for the indicated time. Cell counts normalized to 2d-ctr. (b) Control (ctr), RIN1 knockdown (RIN1 kd) and knockdown rescued with mouse Rin1 (RIN1 kd+mRin1) TOM-1 cells were cultured in 8 μ imatinib for 9 days. (c) Control, RIN1 kd and RIN1 kd+mRin1 TOM-1 cells were immunoblotted with anti-phosphotyrosine. TUBB and RIN1 immunoblots are shown below. Murine Rin1 and human RIN1 were detected using different antibodies. Note: hRIN1 bands are from the same exposure of a single immunoblot. (d) Control (ctr) and RIN1-silenced (kd) B-ALL cells were cultured in 10 μ imatinib for the indicated time. Cell viability was determined by propidium iodide stain and flow cytometry. Panels a, b and d: s.d. from triplicate samples counted in duplicate; * indicates P<0.05 between control and knockdown.
Figure 6
Model of RIN1 effect on BCR-ABL1 activity and imatinib sensitivity. BCR-ABL1 equilibrates between active and inactive conformations, favoring the active form (open to substrate (S)) relative to ABL1. Right: RIN1 binds to ABL1 SH3 and SH2 domains, alleviating residual autoinhibition and stabilizing a high activity conformation. RIN1 is also a RAS effector., Left: Imatinib (IM) preferentially binds and stabilizes the inactive conformation of the ABL1 catalytic site. RIN1 overexpression shifts equilibrium to right; RIN1 silencing shifts equilibrium to left.
Similar articles
- ABL SH3 mutant inhibits BCR-ABL activity and increases imatinib sensitivity by targeting RIN1 protein in CML cell.
Liu X, Li Y, Wen L, Tao K, Xiao Q, Cao W, Huang Z, Gao M, Li H, Wang F, Feng W. Liu X, et al. Cancer Lett. 2015 Dec 1;369(1):222-8. doi: 10.1016/j.canlet.2015.08.017. Epub 2015 Aug 28. Cancer Lett. 2015. PMID: 26321052 - Fusion of EML1 to ABL1 in T-cell acute lymphoblastic leukemia with cryptic t(9;14)(q34;q32).
De Keersmaecker K, Graux C, Odero MD, Mentens N, Somers R, Maertens J, Wlodarska I, Vandenberghe P, Hagemeijer A, Marynen P, Cools J. De Keersmaecker K, et al. Blood. 2005 Jun 15;105(12):4849-52. doi: 10.1182/blood-2004-12-4897. Epub 2005 Feb 15. Blood. 2005. PMID: 15713800 - Dihydroartemisinin inhibits the Bcr/Abl oncogene at the mRNA level in chronic myeloid leukemia sensitive or resistant to imatinib.
Lee J, Shen P, Zhang G, Wu X, Zhang X. Lee J, et al. Biomed Pharmacother. 2013 Mar;67(2):157-63. doi: 10.1016/j.biopha.2012.10.017. Epub 2012 Nov 19. Biomed Pharmacother. 2013. PMID: 23201011 - Mutated tyrosine kinases as therapeutic targets in myeloid leukemias.
Sattler M, Scheijen B, Weisberg E, Griffin JD. Sattler M, et al. Adv Exp Med Biol. 2003;532:121-40. doi: 10.1007/978-1-4615-0081-0_11. Adv Exp Med Biol. 2003. PMID: 12908554 Review. - New Bcr-Abl inhibitors in chronic myeloid leukemia: keeping resistance in check.
O'Hare T, Eide CA, Deininger MW. O'Hare T, et al. Expert Opin Investig Drugs. 2008 Jun;17(6):865-78. doi: 10.1517/13543784.17.6.865. Expert Opin Investig Drugs. 2008. PMID: 18491988 Review.
Cited by
- RIN1 orchestrates the activation of RAB5 GTPases and ABL tyrosine kinases to determine the fate of EGFR.
Balaji K, Mooser C, Janson CM, Bliss JM, Hojjat H, Colicelli J. Balaji K, et al. J Cell Sci. 2012 Dec 1;125(Pt 23):5887-96. doi: 10.1242/jcs.113688. Epub 2012 Sep 12. J Cell Sci. 2012. PMID: 22976291 Free PMC article. - Changing the subcellular location of the oncoprotein Bcr-Abl using rationally designed capture motifs.
Dixon AS, Constance JE, Tanaka T, Rabbitts TH, Lim CS. Dixon AS, et al. Pharm Res. 2012 Apr;29(4):1098-109. doi: 10.1007/s11095-011-0654-8. Epub 2011 Dec 20. Pharm Res. 2012. PMID: 22183511 Free PMC article. - Development of an in vitro genotoxicity assay to detect retroviral vector-induced lymphoid insertional mutants.
Bastone AL, Dziadek V, John-Neek P, Mansel F, Fleischauer J, Agyeman-Duah E, Schaudien D, Dittrich-Breiholz O, Schwarzer A, Schambach A, Rothe M. Bastone AL, et al. Mol Ther Methods Clin Dev. 2023 Aug 22;30:515-533. doi: 10.1016/j.omtm.2023.08.017. eCollection 2023 Sep 14. Mol Ther Methods Clin Dev. 2023. PMID: 37693949 Free PMC article. - The RAB5-GEF function of RIN1 regulates multiple steps during Listeria monocytogenes infection.
Balaji K, French CT, Miller JF, Colicelli J. Balaji K, et al. Traffic. 2014 Nov;15(11):1206-18. doi: 10.1111/tra.12204. Epub 2014 Sep 4. Traffic. 2014. PMID: 25082076 Free PMC article. - Global phosphoproteomics reveals crosstalk between Bcr-Abl and negative feedback mechanisms controlling Src signaling.
Rubbi L, Titz B, Brown L, Galvan E, Komisopoulou E, Chen SS, Low T, Tahmasian M, Skaggs B, Müschen M, Pellegrini M, Graeber TG. Rubbi L, et al. Sci Signal. 2011 Mar 29;4(166):ra18. doi: 10.1126/scisignal.2001314. Sci Signal. 2011. PMID: 21447799 Free PMC article.
References
- Wong S, Witte ON. The BCR-ABL story: bench to bedside and back. Annu Rev Immunol. 2004;22:247–306. - PubMed
- Papadopoulos P, Ridge SA, Boucher CA, Stocking C, Wiedemann LM. The novel activation of ABL by fusion to an ets-related gene, TEL. Cancer Res. 1995;55:34–38. - PubMed
- Iijima Y, Ito T, Oikawa T, Eguchi M, Eguchi-Ishimae M, Kamada N, et al. A new ETV6/TEL partner gene, ARG (ABL-related gene or ABL2), identified in an AML-M3 cell line with a t(1;12)(q25;p13) translocation. Blood. 2000;95:2126–2131. - PubMed
- Deininger M, Buchdunger E, Druker BJ. The development of imatinib as a therapeutic agent for chronic myeloid leukemia. Blood. 2005;105:2640–2653. - PubMed
- Gorre ME, Mohammed M, Ellwood K, Hsu N, Paquette R, Rao PN, et al. Clinical resistance to STI-571 cancer therapy caused by BCR-ABL gene mutation or amplification. Science (NY) 2001;293:876–880. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 CA136699/CA/NCI NIH HHS/United States
- R01 NS046787/NS/NINDS NIH HHS/United States
- HHMI/Howard Hughes Medical Institute/United States
- CA136699/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous