Characterization of the fecal microbiome from non-human wild primates reveals species specific microbial communities - PubMed (original) (raw)
Characterization of the fecal microbiome from non-human wild primates reveals species specific microbial communities
Suleyman Yildirim et al. PLoS One. 2010.
Abstract
Background: Host-associated microbes comprise an integral part of animal digestive systems and these interactions have a long evolutionary history. It has been hypothesized that the gastrointestinal microbiome of humans and other non-human primates may have played significant roles in host evolution by facilitating a range of dietary adaptations. We have undertaken a comparative sequencing survey of the gastrointestinal microbiomes of several non-human primate species, with the goal of better understanding how these microbiomes relate to the evolution of non-human primate diversity. Here we present a comparative analysis of gastrointestinal microbial communities from three different species of Old World wild monkeys.
Methodology/principal findings: We analyzed fecal samples from three different wild non-human primate species (black-and-white colobus [Colubus guereza], red colobus [Piliocolobus tephrosceles], and red-tailed guenon [Cercopithecus ascanius]). Three samples from each species were subjected to small subunit rRNA tag pyrosequencing. Firmicutes comprised the vast majority of the phyla in each sample. Other phyla represented were Bacterioidetes, Proteobacteria, Spirochaetes, Actinobacteria, Verrucomicrobia, Lentisphaerae, Tenericutes, Planctomycetes, Fibrobacateres, and TM7. Bray-Curtis similarity analysis of these microbiomes indicated that microbial community composition within the same primate species are more similar to each other than to those of different primate species. Comparison of fecal microbiota from non-human primates with microbiota of human stool samples obtained in previous studies revealed that the gut microbiota of these primates are distinct and reflect host phylogeny.
Conclusion/significance: Our analysis provides evidence that the fecal microbiomes of wild primates co-vary with their hosts, and that this is manifested in higher intraspecies similarity among wild primate species, perhaps reflecting species specificity of the microbiome in addition to dietary influences. These results contribute to the limited body of primate microbiome studies and provide a framework for comparative microbiome analysis between human and non-human primates as well as a comparative evolutionary understanding of the human microbiome.
Conflict of interest statement
Competing Interests: The authors have declared that no competing interests exist.
Figures
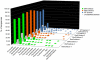
Figure 1. Relative abundance of phylum members of fecal bacteria from nine wild primate subjects.
Ribosomal Database Project classifier (v.10.2; 70% confidence threshold) was used for sequence assignment. Sequence abundance from fecal samples of black-and-white colobus subjects are green; red colobus subjects are red; red-tailed guenon subjects are blue, and unclassified bacteria abundance are represented by grey color bars. The analysis is based on 534R dataset.
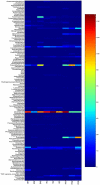
Figure 2. Heatmap display of the relative abundance of genera.
(534R dataset). The color spectrum represents abundance of each genus per thousand sequences; dark blue representing minimum relative abundance and dark red representing maximum relative abundance. The genera are shown in alphabetical order. Each subject is shown at the bottom of the figure; BW1-BW3: Black and white colobus subjects; RC1-RC3; Red colobus subjects, and RTG1-RTG3: Red-tailed guenon subjects.
Figure 3. Multi Dimensional Scale analysis of the microbiomes based on the 534R dataset.
non-metric Multi Dimensional Scale analysis is an ordination method used to visually assess variation in patterns of microbial diversity. Each data point represents 16S rRNA data from a single primate subject; Black and white colobus subjects, red-colobus subjects, and red-tailed guenon subjects are designated by symbols in green, red, and blue colors, respectively. The Bray-Curtis similarity was used to rank the distances calculated using the community data.
Figure 4. Multi Dimensional Scale analysis of the microbiomes based on the 27F dataset.
Each data point represents 16S rRNA data from a single primate subject; Black and white colobus subjects, red-colobus subjects, and red-tailed guenon subjects are designated by symbols in green, red, and blue colors, respectively. The Bray-Curtis similarity was used to rank the distances calculated using the community data.
Figure 5. Multi Dimensional Scale analysis of the microbiomes based on the (27F-534R) dataset.
Black and white colobus subjects, red-colobus subjects, and red-tailed guenon subjects are designated by symbols in green, red, and blue colors, respectively. The Bray-Curtis similarity was used to rank the distances calculated using the community data.
Figure 6. Multi Dimensional Scale analysis of the microbiomes based on the enriched (27F-534R) dataset.
The analysis was based on the (27F-534R) dataset, which include human stool data. Human subjects (A, B, C) were previously described elsewhere . Black and white colobus subjects, red-colobus subjects, red-tailed guenon subjects, and human subjects are designated by symbols in green, orange, blue, and purple colors, respectively. The Bray-Curtis similarity was used to rank the distances calculated using the community data.
Similar articles
- Fecal microbiomes of non-human primates in Western Uganda reveal species-specific communities largely resistant to habitat perturbation.
McCord AI, Chapman CA, Weny G, Tumukunde A, Hyeroba D, Klotz K, Koblings AS, Mbora DN, Cregger M, White BA, Leigh SR, Goldberg TL. McCord AI, et al. Am J Primatol. 2014 Apr;76(4):347-54. doi: 10.1002/ajp.22238. Epub 2013 Nov 27. Am J Primatol. 2014. PMID: 24285224 Free PMC article. - Plasticity in the Human Gut Microbiome Defies Evolutionary Constraints.
Gomez A, Sharma AK, Mallott EK, Petrzelkova KJ, Jost Robinson CA, Yeoman CJ, Carbonero F, Pafco B, Rothman JM, Ulanov A, Vlckova K, Amato KR, Schnorr SL, Dominy NJ, Modry D, Todd A, Torralba M, Nelson KE, Burns MB, Blekhman R, Remis M, Stumpf RM, Wilson BA, Gaskins HR, Garber PA, White BA, Leigh SR. Gomez A, et al. mSphere. 2019 Jul 31;4(4):e00271-19. doi: 10.1128/mSphere.00271-19. mSphere. 2019. PMID: 31366708 Free PMC article. - Large Comparative Analyses of Primate Body Site Microbiomes Indicate that the Oral Microbiome Is Unique among All Body Sites and Conserved among Nonhuman Primates.
Asangba AE, Mugisha L, Rukundo J, Lewis RJ, Halajian A, Cortés-Ortiz L, Junge RE, Irwin MT, Karlson J, Perkin A, Watsa M, Erkenswick G, Bales KL, Patton DL, Jasinska AJ, Fernandez-Duque E, Leigh SR, Stumpf RM. Asangba AE, et al. Microbiol Spectr. 2022 Jun 29;10(3):e0164321. doi: 10.1128/spectrum.01643-21. Epub 2022 May 19. Microbiol Spectr. 2022. PMID: 35587638 Free PMC article. - Worlds within worlds: evolution of the vertebrate gut microbiota.
Ley RE, Lozupone CA, Hamady M, Knight R, Gordon JI. Ley RE, et al. Nat Rev Microbiol. 2008 Oct;6(10):776-88. doi: 10.1038/nrmicro1978. Nat Rev Microbiol. 2008. PMID: 18794915 Free PMC article. Review. - The gut microbiome of nonhuman primates: Lessons in ecology and evolution.
Clayton JB, Gomez A, Amato K, Knights D, Travis DA, Blekhman R, Knight R, Leigh S, Stumpf R, Wolf T, Glander KE, Cabana F, Johnson TJ. Clayton JB, et al. Am J Primatol. 2018 Jun;80(6):e22867. doi: 10.1002/ajp.22867. Epub 2018 Jun 4. Am J Primatol. 2018. PMID: 29862519 Review.
Cited by
- Microbes Drive Evolution of Animals and Plants: the Hologenome Concept.
Rosenberg E, Zilber-Rosenberg I. Rosenberg E, et al. mBio. 2016 Mar 31;7(2):e01395. doi: 10.1128/mBio.01395-15. mBio. 2016. PMID: 27034283 Free PMC article. Review. - Host phylogeny and life history stage shape the gut microbiome in dwarf (Kogia sima) and pygmy (Kogia breviceps) sperm whales.
Denison ER, Rhodes RG, McLellan WA, Pabst DA, Erwin PM. Denison ER, et al. Sci Rep. 2020 Sep 16;10(1):15162. doi: 10.1038/s41598-020-72032-4. Sci Rep. 2020. PMID: 32938948 Free PMC article. - Primate microbiomes over time: Longitudinal answers to standing questions in microbiome research.
Björk JR, Dasari M, Grieneisen L, Archie EA. Björk JR, et al. Am J Primatol. 2019 Oct;81(10-11):e22970. doi: 10.1002/ajp.22970. Epub 2019 Apr 2. Am J Primatol. 2019. PMID: 30941803 Free PMC article. Review. - Age-related changes in the marmoset gut microbiome.
Reveles KR, Patel S, Forney L, Ross CN. Reveles KR, et al. Am J Primatol. 2019 Feb;81(2):e22960. doi: 10.1002/ajp.22960. Epub 2019 Feb 25. Am J Primatol. 2019. PMID: 30802990 Free PMC article. - Effects of green tea on miRNA and microbiome of oral epithelium.
Adami GR, Tangney CC, Tang JL, Zhou Y, Ghaffari S, Naqib A, Sinha S, Green SJ, Schwartz JL. Adami GR, et al. Sci Rep. 2018 Apr 12;8(1):5873. doi: 10.1038/s41598-018-22994-3. Sci Rep. 2018. PMID: 29651001 Free PMC article.
References
- Dale C, Moran NA. Molecular interactions between bacterial symbionts and their hosts. Cell. 2006;126:453–65. - PubMed
- Hickman C. How bacteria contributed to the evolution of multicellular animals? In: McFall Ngai MargaretJ., editor. The Influence of Cooperative Bacteria on Animal Host Biology. New York: Cambridge University Press; 2005. pp. 3–35.
- Hooper LV, Gordon JI. Commensal host-bacterial relationships in the gut. Science. 2001;292:1115–8. - PubMed
- Fleagle JG. London: Academic Press; 1999. Primate Adaptation and Evolution.291 2nd Edition.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical