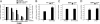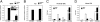Nicotinamide adenine dinucleotide (NAD)-regulated DNA methylation alters CCCTC-binding factor (CTCF)/cohesin binding and transcription at the BDNF locus - PubMed (original) (raw)
Nicotinamide adenine dinucleotide (NAD)-regulated DNA methylation alters CCCTC-binding factor (CTCF)/cohesin binding and transcription at the BDNF locus
Jufang Chang et al. Proc Natl Acad Sci U S A. 2010.
Abstract
Cellular metabolism alters patterns of gene expression through a variety of mechanisms, including alterations in histone modifications and transcription factor activity. Nicotinamide adenine dinucleotide (NAD)-dependent proteins such as poly(ADP ribose) polymerases (PARPs) and sirtuin deacetylases play important roles in this regulation, thus NAD provides a crucial link between metabolism and these cellular signaling processes. Here, we found that lowering NAD levels in mouse primary cortical neurons led to decreased activity-dependent BDNF expression. The altered BDNF transcription occurred independently of Sirt or Parp activities; instead, low NAD levels promoted increased DNA methylation of the activity-dependent BDNF promoter. Increased methylation at this promoter triggered the dissociation of the insulator protein CTCF as well as the accompanying cohesin from the BDNF locus. The loss of these proteins resulted in histone acetylation and methylation changes at this locus consistent with chromatin compaction and gene silencing. Because BDNF is critical for neuronal function, these results suggest that age- or nutrition-associated declines in NAD levels as well as deficits in cohesin function associated with disease modulate BDNF expression and could contribute to cognitive impairment.
Conflict of interest statement
Conflict of interest statement: The authors and Washington University may derive benefit from a licensing agreement with Sirtris Pharmaceuticals, which did not provide any support for this work.
Figures
Fig. 1.
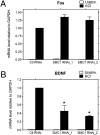
NAD levels regulate activity-dependent BDNF transcription in cortical neurons. BDNF transcripts initiated from promoter I, II, IV, and VI, as well as the level of total BDNF transcripts, were measured by using qRT-PCR and normalized to GAPDH mRNA levels. Mouse cortical neurons were stimulated with KCl for 4 h to induce membrane depolarization and BDNF transcription. Neurons treated with Nampt inhibitor FK866 (10 nM) had decreased BDNF induction after KCl treatment.
Fig. 2.
Activity-dependent BDNF transcription initiated from promoter IV in cortical neurons is inhibited by FK866. (A) NAD levels and transcription from BDNF promoter IV in mouse cortical neurons treated with indicated concentration of the Nampt inhibitor FK866. Nicotinamide mononucleotide (NMN), the product of the Nampt-catalyzed reaction, was added where indicated. Total mRNA was isolated from naïve neurons or those stimulated with KCl for 4 h. BDNF transcripts initiated from promoter IV were measured with qRT-PCR and normalized to GAPDH mRNA levels. Neurons with low NAD levels had decreased BDNF induction after KCl treatment. (B) c-fos mRNA levels in neurons incubated with or without FK866 were determined as above and were not affected by FK866 treatment. (C) E15 primary cortical neurons from wild-type and Sirt1-deficient mice were cultured for 5DIV and stimulated with 55 mM KCl for 4 h. Sirt1-deficient neurons showed equivalent BDNF induction compared with wild-type controls. Quantification was performed from duplicate experiments from at least three mice for each genotype. (D) Mouse cortical neurons were incubated for 24 h with PARP inhibitor 3-aminobenzamide (3-ABA; 7 mM) before depolarization by KCl addition. Inhibition of PARPs had no impact on the magnitude of BDNF induction. Quantification was performed from three independent experiments.
Fig. 3.
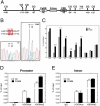
FK866-treated cortical neurons show increased levels of DNA methylation at BDNF promoter IV. (A) A schematic depicting the regulatory elements within the mouse BDNF exon IV promoter (promoter IV) (6). (B) Direct sequencing of PCR products amplified with bisulfate-treated DNA as template. DNA samples were derived from 5DIV cortical neurons treated with or without FK866. DNA sequencing traces are shown for the −109 and −111 sites in BDNF promoter IV. Note the traces depict the antisense (reverse) strand; therefore, G is indicative of a methylated cytosine, and A is indicative of a nonmethylated cytosine. An increase in DNA methylation at −109 and −111 sites was observed in samples treated with FK866. (C) Mapping of the methylation status of the 10 CpG sites near the transcription start site (TSS) by bisulfate sequencing of genomic DNA. The number above the black bar represents the fold changes in methylated cytosine relative to control at the indicated position. (D and E) Neurons treated with or without FK866 (10 nM) for 24 h were subjected to ChIP with antibodies specific to trimethyl-H3 K9 (H3K9Me3), acetyl-H3 K9 (H3K9Ace), or control IgG followed by amplification with PCR primers specific for BDNF promoter IV and a downstream intronic region. Data show the representative result from four independent experiments.
Fig. 4.
CTCF and cohesin are associated with the BDNF locus in mouse cortical neurons. (A) Previously published (20) whole-genome ChIP data was visualized with the Integrated Genome Browser (Affymetrix) to identify CTCF binding sites at the BDNF locus. CTCF binds to multiple sites on BDNF locus in HeLa cells, and the numbers under the peaks indicate order of peak magnitude in BDNF locus; for example, 1 is the site with the highest binding and 8 is the lowest. (B) A schematic diagram illustrating the genomic structure of the mouse BDNF gene (4). Solid boxes represent exons, and encircled numbers indicate positions of CTCF binding sites labeled above relative to these exons. (C and D) ChIP assays using antibodies specific to CTCF or the core cohesin component SCC1 were performed to assess occupancy of indicated sites within BDNF locus in cortical neurons. CTCF and cohesin were enriched at BDNF promoter IV (P5) and at P1 in the intron. Fragments corresponding to N1 and N2 were used as negative control binding sites for CTCF and SCC1. Data show the representative result from six independent experiments.
Fig. 5.
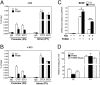
CTCF and cohesin binding to BDNF promoter IV is regulated by NAD via altered DNA methylation. (A and B) Cortical neurons treated with or without FK866 for 24 h (and with or without KCl for 90 min) were subjected to ChIP analysis with antibodies specific to CTCF or SCC1. Low NAD levels promoted the release of CTCF and SCC1 from the BDNF promoter IV (P5) but not from the intronic region (P1). (C) Restoration of BDNF gene expression after 5AzaC pretreatment of cortical neurons. KCl-induced BDNF transcription initiated at promoter IV was measured by qRT-PCR in neurons treated with FK866 in the absence (−) or presence (+) of 5AzaC. Results are mean ± SD. *P < 0.01, **P < 0.0001. Data are representative of at least four independent experiments. (D) ChIP analysis showed increased levels of CTCF occupancy at BDNF promoter IV (P5) but not at the intron (P1) in the presence of FK866 in neurons pretreated with 5AzaC.
Fig. 6.
CTCF and cohesin proteins are essential for BDNF transcription in mouse cortical neurons. (A) Western blot analysis of lysates prepared from CTCFf/f cortical neurons infected with lentivirus expressing EGFP or Cre recombinase. Note the complete loss of CTCF in neurons expressing Cre, indicating efficient excision of the mutant allele (Insert). BDNF transcripts initiated at promoter IV in control or CTCF-depleted neurons were measured by qRT-PCR. Neurons lacking CTCF had significantly decreased levels of BDNF mRNA after KCl treatment. Results are mean ± SD. *P < 0.0001 (vs. F/F_GFP treated with KCl). The results shown are representative of those obtained in five independent experiments. (B) RT-PCR showing normal levels of c-fos mRNA in KCl-stimulated cortical neurons lacking CTCF. (C and D) ChIP analysis of CTCF, SCC1, and H3K9Me3 (inactive chromatin marker) binding at BDNF promoter IV (P5) and intronic region (P1) in CTCFf/f cortical neurons infected with lentivirus expressing EGFP or Cre recombinase. Note that loss of CTCF resulted in loss of SCC1 at these sites and generated an inactive chromatin structure in this region.
Fig. 7.
Neurons lacking SMC1 have defects in activity-induced BDNF transcription. Cortical neurons were infected with lentivirus expressing siRNAs targeting luciferase (control) or SMC1 and stimulated with KCl for 4 h. c-fos (A) and BDNF (B) mRNA levels were measured by using qRT-PCR. Neurons depleted of SMC1 had normal KCl-mediated induction of c-fos but BDNF induction was greatly decreased, similar to that observed in neurons lacking CTCF. The error bars indicate the ± SD. *P < 0.0001 (vs. Ctl RNAi treated with KCl). Data represent at least three independent experiments.
Similar articles
- Disruption of CTCF/cohesin-mediated high-order chromatin structures by DNA methylation downregulates PTGS2 expression.
Kang JY, Song SH, Yun J, Jeon MS, Kim HP, Han SW, Kim TY. Kang JY, et al. Oncogene. 2015 Nov 5;34(45):5677-84. doi: 10.1038/onc.2015.17. Epub 2015 Feb 23. Oncogene. 2015. PMID: 25703332 - CCCTC-binding factor (CTCF) and cohesin influence the genomic architecture of the Igh locus and antisense transcription in pro-B cells.
Degner SC, Verma-Gaur J, Wong TP, Bossen C, Iverson GM, Torkamani A, Vettermann C, Lin YC, Ju Z, Schulz D, Murre CS, Birshtein BK, Schork NJ, Schlissel MS, Riblet R, Murre C, Feeney AJ. Degner SC, et al. Proc Natl Acad Sci U S A. 2011 Jun 7;108(23):9566-71. doi: 10.1073/pnas.1019391108. Epub 2011 May 23. Proc Natl Acad Sci U S A. 2011. PMID: 21606361 Free PMC article. - Genome-wide and parental allele-specific analysis of CTCF and cohesin DNA binding in mouse brain reveals a tissue-specific binding pattern and an association with imprinted differentially methylated regions.
Prickett AR, Barkas N, McCole RB, Hughes S, Amante SM, Schulz R, Oakey RJ. Prickett AR, et al. Genome Res. 2013 Oct;23(10):1624-35. doi: 10.1101/gr.150136.112. Epub 2013 Jun 26. Genome Res. 2013. PMID: 23804403 Free PMC article. - Genome-wide studies of CCCTC-binding factor (CTCF) and cohesin provide insight into chromatin structure and regulation.
Lee BK, Iyer VR. Lee BK, et al. J Biol Chem. 2012 Sep 7;287(37):30906-13. doi: 10.1074/jbc.R111.324962. Epub 2012 Sep 5. J Biol Chem. 2012. PMID: 22952237 Free PMC article. Review. - How cohesin and CTCF cooperate in regulating gene expression.
Wendt KS, Peters JM. Wendt KS, et al. Chromosome Res. 2009;17(2):201-14. doi: 10.1007/s10577-008-9017-7. Epub 2009 Mar 24. Chromosome Res. 2009. PMID: 19308701 Review.
Cited by
- A randomized placebo-controlled trial of nicotinamide riboside in older adults with mild cognitive impairment.
Orr ME, Kotkowski E, Ramirez P, Bair-Kelps D, Liu Q, Brenner C, Schmidt MS, Fox PT, Larbi A, Tan C, Wong G, Gelfond J, Frost B, Espinoza S, Musi N, Powers B. Orr ME, et al. Geroscience. 2024 Feb;46(1):665-682. doi: 10.1007/s11357-023-00999-9. Epub 2023 Nov 23. Geroscience. 2024. PMID: 37994989 Free PMC article. Clinical Trial. - Active DNA demethylation in post-mitotic neurons: a reason for optimism.
Gavin DP, Chase KA, Sharma RP. Gavin DP, et al. Neuropharmacology. 2013 Dec;75:233-45. doi: 10.1016/j.neuropharm.2013.07.036. Epub 2013 Aug 16. Neuropharmacology. 2013. PMID: 23958448 Free PMC article. Review. - Role of primary aging hallmarks in Alzheimer´s disease.
Zhao J, Huai J. Zhao J, et al. Theranostics. 2023 Jan 1;13(1):197-230. doi: 10.7150/thno.79535. eCollection 2023. Theranostics. 2023. PMID: 36593969 Free PMC article. Review. - Neuron-specific impairment of inter-chromosomal pairing and transcription in a novel model of human 15q-duplication syndrome.
Meguro-Horike M, Yasui DH, Powell W, Schroeder DI, Oshimura M, Lasalle JM, Horike S. Meguro-Horike M, et al. Hum Mol Genet. 2011 Oct 1;20(19):3798-810. doi: 10.1093/hmg/ddr298. Epub 2011 Jul 1. Hum Mol Genet. 2011. PMID: 21725066 Free PMC article. - An NAD+-dependent transcriptional program governs self-renewal and radiation resistance in glioblastoma.
Gujar AD, Le S, Mao DD, Dadey DY, Turski A, Sasaki Y, Aum D, Luo J, Dahiya S, Yuan L, Rich KM, Milbrandt J, Hallahan DE, Yano H, Tran DD, Kim AH. Gujar AD, et al. Proc Natl Acad Sci U S A. 2016 Dec 20;113(51):E8247-E8256. doi: 10.1073/pnas.1610921114. Epub 2016 Dec 7. Proc Natl Acad Sci U S A. 2016. PMID: 27930300 Free PMC article.
References
- Zuccato C, Cattaneo E. Brain-derived neurotrophic factor in neurodegenerative diseases. Nat Rev Neurol. 2009;5:311–322. - PubMed
- West AE. Biological functions of activity-dependent transcription revealed. Neuron. 2008;60:523–525. - PubMed
- Timmusk T, et al. Multiple promoters direct tissue-specific expression of the rat BDNF gene. Neuron. 1993;10:475–489. - PubMed
- Chen WG, et al. Derepression of BDNF transcription involves calcium-dependent phosphorylation of MeCP2. Science. 2003;302:885–889. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- P30 NS057105/NS/NINDS NIH HHS/United States
- AG013730/AG/NIA NIH HHS/United States
- R01 AG013730/AG/NIA NIH HHS/United States
- CA111966/CA/NCI NIH HHS/United States
- R01 CA111966/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases