Genome sequence of adherent-invasive Escherichia coli and comparative genomic analysis with other E. coli pathotypes - PubMed (original) (raw)
Comparative Study
Genome sequence of adherent-invasive Escherichia coli and comparative genomic analysis with other E. coli pathotypes
John He Nash et al. BMC Genomics. 2010.
Abstract
Background: Adherent and invasive Escherichia coli (AIEC) are commonly found in ileal lesions of Crohn's Disease (CD) patients, where they adhere to intestinal epithelial cells and invade into and survive in epithelial cells and macrophages, thereby gaining access to a typically restricted host niche. Colonization leads to strong inflammatory responses in the gut suggesting that AIEC could play a role in CD immunopathology. Despite extensive investigation, the genetic determinants accounting for the AIEC phenotype remain poorly defined. To address this, we present the complete genome sequence of an AIEC, revealing the genetic blueprint for this disease-associated E. coli pathotype.
Results: We sequenced the complete genome of E. coli NRG857c (O83:H1), a clinical isolate of AIEC from the ileum of a Crohn's Disease patient. Our sequence data confirmed a phylogenetic linkage between AIEC and extraintestinal pathogenic E. coli causing urinary tract infections and neonatal meningitis. The comparison of the NRG857c AIEC genome with other pathogenic and commensal E. coli allowed for the identification of unique genetic features of the AIEC pathotype, including 41 genomic islands, and unique genes that are found only in strains exhibiting the adherent and invasive phenotype.
Conclusions: Up to now, the virulence-like features associated with AIEC are detectable only phenotypically. AIEC genome sequence data will facilitate the identification of genetic determinants implicated in invasion and intracellular growth, as well as enable functional genomic studies of AIEC gene expression during health and disease.
Figures
Figure 1
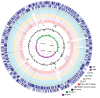
Comparative genome atlas of NRG857c. The chromosome of NRG857c (two outermost rings are CDS on forward and reverse strand) was compared with those of selected E. coli strains, starting from the outer layer LF82 (AIEC; pale green), APEC-O1 (APEC; blue), CFT073 (UPEC; yellow), MG1655 (K12/commensal; purple) and enterohemorrhagic E. coli O157:H7 Sakai (EHEC, red). Genomic islands were plotted on the NRG857c chromosome (grey blocks). The G+C content and G/C skew are also plotted as indicated.
Figure 2
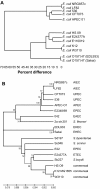
Phylogenetic analysis of NRG857c compared with representative strains of other enteric bacteria. (A) A phylogenetic tree based on the unweighted pair group method with arithmetic mean was constructed from the optimal map data and in silico Nco_I_ restriction digests of other enteric bacterial chromosomes. (B) MLST-based analysis of NRG857c with other enteric bacteria was performed as described in the Methods and sequence data was used to construct a phylogenetic tree. Numbers on the tree branches represent bootstrap support from 1000 bootstrap replicates with a minimum cut-off of 65%. Accession numbers for gene sequences can be found in Additional File 2, Table S1.
Figure 3
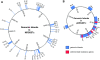
Genomic islands in NRG857c. Genomic islands in the NRG857c chromosome (A) and plasmid (B) were predicted using stringent bioinformatics criteria as described in the Methods. Genomic islands are plotted to scale in blue and labelled clockwise on the genome maps. On the plasmid, genes involved in antimicrobial resistance are indicated in red.
Figure 4
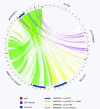
Gene content analysis of plasmid pNRG857c and comparison to representative strains of other E. coli. BLASTN analysis was performed between each CDS in plasmid pNRG857c against each CDS in pLF82, pO157Sakai, pUTI89, and pAPEC-O1-ColBM. Genes in pNRG857c with orthologs in the other plasmids, defined as >85% identity over entire length of the gene, are connected with a coloured line.
Figure 5
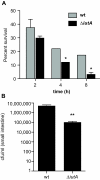
Iron uptake by the aerobactin system is important for intracellular survival and for mouse colonization. (A) J774.A1 macrophage cells were infected with wild type NRG857c or iutA mutant cells. The survival of intracellular bacteria was determined at various times after infection. Data are the mean survival of intracellular bacteria with standard deviation. (*, P < 0.05, Mann Whitney) (B) The aerobactin iron transport system improves colonization in vivo. Groups of mice were infected orally with wild type NRG857c or iutA mutants. Colonization of the small intestine by NRG857c AIEC was determined three days after infection by enumerating the number of cfu in tissue homogenates. Data are the means with standard errors. (**, P < 0.005, Mann Whitney).
Similar articles
- Comparative genomics of a subset of Adherent/Invasive Escherichia coli strains isolated from individuals without inflammatory bowel disease.
Barrios-Villa E, Martínez de la Peña CF, Lozano-Zaraín P, Cevallos MA, Torres C, Torres AG, Rocha-Gracia RDC. Barrios-Villa E, et al. Genomics. 2020 Mar;112(2):1813-1820. doi: 10.1016/j.ygeno.2019.10.013. Epub 2019 Nov 2. Genomics. 2020. PMID: 31689478 - Similarity and divergence among adherent-invasive Escherichia coli and extraintestinal pathogenic E. coli strains.
Martinez-Medina M, Mora A, Blanco M, López C, Alonso MP, Bonacorsi S, Nicolas-Chanoine MH, Darfeuille-Michaud A, Garcia-Gil J, Blanco J. Martinez-Medina M, et al. J Clin Microbiol. 2009 Dec;47(12):3968-79. doi: 10.1128/JCM.01484-09. Epub 2009 Oct 14. J Clin Microbiol. 2009. PMID: 19828750 Free PMC article. - Comparative genomics reveals new single-nucleotide polymorphisms that can assist in identification of adherent-invasive Escherichia coli.
Camprubí-Font C, Lopez-Siles M, Ferrer-Guixeras M, Niubó-Carulla L, Abellà-Ametller C, Garcia-Gil LJ, Martinez-Medina M. Camprubí-Font C, et al. Sci Rep. 2018 Feb 9;8(1):2695. doi: 10.1038/s41598-018-20843-x. Sci Rep. 2018. PMID: 29426864 Free PMC article. - The Unique Lifestyle of Crohn's Disease-Associated Adherent-Invasive Escherichia coli.
Shaler CR, Elhenawy W, Coombes BK. Shaler CR, et al. J Mol Biol. 2019 Jul 26;431(16):2970-2981. doi: 10.1016/j.jmb.2019.04.023. Epub 2019 Apr 25. J Mol Biol. 2019. PMID: 31029703 Review. - Pathogenesis of adherent-invasive Escherichia coli.
Smith EJ, Thompson AP, O'Driscoll A, Clarke DJ. Smith EJ, et al. Future Microbiol. 2013 Oct;8(10):1289-300. doi: 10.2217/fmb.13.94. Future Microbiol. 2013. PMID: 24059919 Review.
Cited by
- Characterization of the genetic environment of bla ESBL genes, integrons and toxin-antitoxin systems identified on large transferrable plasmids in multi-drug resistant Escherichia coli.
Wang J, Stephan R, Zurfluh K, Hächler H, Fanning S. Wang J, et al. Front Microbiol. 2015 Jan 6;5:716. doi: 10.3389/fmicb.2014.00716. eCollection 2014. Front Microbiol. 2015. PMID: 25610429 Free PMC article. - Epidemiology of E. coli in Cystic Fibrosis Airways Demonstrates the Capacity for Persistent Infection but Not Patient-Patient Transmission.
Izydorczyk C, Waddell B, Edwards BD, Greysson-Wong J, Surette MG, Somayaji R, Rabin HR, Conly JM, Church DL, Parkins MD. Izydorczyk C, et al. Front Microbiol. 2020 Mar 20;11:475. doi: 10.3389/fmicb.2020.00475. eCollection 2020. Front Microbiol. 2020. PMID: 32265892 Free PMC article. - Adaptations of Escherichia coli strains to oxidative stress are reflected in properties of their structural proteomes.
Mih N, Monk JM, Fang X, Catoiu E, Heckmann D, Yang L, Palsson BO. Mih N, et al. BMC Bioinformatics. 2020 Apr 29;21(1):162. doi: 10.1186/s12859-020-3505-y. BMC Bioinformatics. 2020. PMID: 32349661 Free PMC article. - Adherent-Invasive and Non-Invasive Escherichia coli Isolates Differ in Their Effects on Caenorhabditis elegans' Lifespan.
de Sousa Figueiredo MB, Pradel E, George F, Mahieux S, Houcke I, Pottier M, Fradin C, Neut C, Daniel C, Bongiovanni A, Foligné B, Titécat M. de Sousa Figueiredo MB, et al. Microorganisms. 2021 Aug 27;9(9):1823. doi: 10.3390/microorganisms9091823. Microorganisms. 2021. PMID: 34576719 Free PMC article. - IBD-what role do Proteobacteria play?
Mukhopadhya I, Hansen R, El-Omar EM, Hold GL. Mukhopadhya I, et al. Nat Rev Gastroenterol Hepatol. 2012 Feb 21;9(4):219-30. doi: 10.1038/nrgastro.2012.14. Nat Rev Gastroenterol Hepatol. 2012. PMID: 22349170 Review.
References
- Sartor RB. Current concepts of the etiology and pathogenesis of ulcerative colitis and Crohn's disease. Gastroenterol Clin North Am. 1995;24:475–507. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases