Initial development and validation of a novel extraction method for quantitative mining of the formalin-fixed, paraffin-embedded tissue proteome for biomarker investigations - PubMed (original) (raw)
. 2011 Feb 4;10(2):896-906.
doi: 10.1021/pr100812d. Epub 2010 Dec 23.
Affiliations
- PMID: 21117664
- PMCID: PMC3033703
- DOI: 10.1021/pr100812d
Free PMC article
Initial development and validation of a novel extraction method for quantitative mining of the formalin-fixed, paraffin-embedded tissue proteome for biomarker investigations
Niroshini J Nirmalan et al. J Proteome Res. 2011.
Free PMC article
Abstract
Annotated formalin-fixed, paraffin-embedded (FFPE) tissue archives constitute a valuable resource for retrospective biomarker discovery. However, proteomic exploration of archival tissue is impeded by extensive formalin-induced covalent cross-linking. Robust methodology enabling proteomic profiling of archival resources is urgently needed. Recent work is beginning to support the feasibility of biomarker discovery in archival tissues, but further developments in extraction methods which are compatible with quantitative approaches are urgently needed. We report a cost-effective extraction methodology permitting quantitative proteomic analyses of small amounts of FFPE tissue for biomarker investigation. This surfactant/heat-based approach results in effective and reproducible protein extraction in FFPE tissue blocks. In combination with a liquid chromatography-mass spectrometry-based label-free quantitative proteomics methodology, the protocol enables the robust representative and quantitative analyses of the archival proteome. Preliminary validation studies in renal cancer tissues have identified typically 250-300 proteins per 500 ng of tissue with 1D LC-MS/MS with comparable extraction in FFPE and fresh frozen tissue blocks and preservation of tumor/normal differential expression patterns (205 proteins, r = 0.682; p < 10(-15)). The initial methodology presented here provides a quantitative approach for assessing the potential suitability of the vast FFPE tissue archives as an alternate resource for biomarker discovery and will allow exploration of methods to increase depth of coverage and investigate the impact of preanalytical factors.
Figures
Figure 1
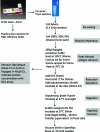
Summary of optimized surfactant/heat-based protein extraction protocol for formalin-fixed, paraffin-embedded tissue blocks.
Figure 2
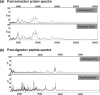
Comparison of representative protein extraction efficiencies for FFPE normal renal tissues with Rapigest and ACN−NH3HCO3 buffers using SELDI-TOF (Ciphergen). Spectral acquisition was carried out on 1 μg protein extracts spotted on NP20 chips. (a) The postextraction protein spectra and (b) the post-tryptic digestion peptide spectra for the two extraction buffers.
Figure 3
Representative example of a comparison of protein extracts (400 ng) from frozen and FFPE extracts in (a) normal renal cortical tissue and (b) RCC tissue, analyzed using a NanoAcquity 1D-UPLC-HDMS quadrupole-oa-Tof (Waters U.K.). Each extract was run in triplicate and proteins appearing in at least 2 of three replicates were included in the analysis.
Figure 4
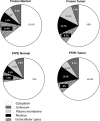
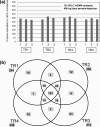
Subcellular localization analyses (Ingenuity IPA software (version 8.0)) of the proteins identified from matched normal renal cortex and RCC tissue extract (FFPE and frozen tissue; Figure 3) using the optimized Rapigest surfactant protocol. The proteins were identified at high confidence using a NanoAcquity UPLC-HDMS quadrupole-oa.Tof with protein loading normalized using a spiked external standard.
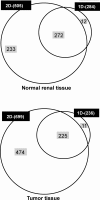
Figure 5
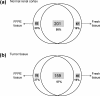
Technical replicate analysis of FFPE normal renal tissue extracts. (a) Proteins identified at high stringency across triplicate injections (1, 2, 3; 400 ng each injection) of each of 4 independently extracted replicates (TR1, TR2, TR3, TR4) from a single normal renal cortex FFPE tissue block; (b) shows the overall distribution patterns of the proteins within the replicate sets analyzed by 1D-LC−MS/MS. A total of 191 proteins were common to all four replicates with TR1, TR2, TR3 and TR4 extracts having 254, 259, 272, and 246 proteins identified, respectively, at high stringency.
Figure 6
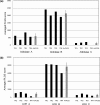
Quantitative analysis of reproducibility of isoform assignment in a series of 4 technical replicates from normal renal cortical FFPE extracts. PLGS scores for the replicates are shown for two selected protein examples, (a) aldolase A, B, C and (b) LDH A, B and C. The quantitation shows averaged PLGS scores from triplicate injections for each protein compared in the 4 technical replicates with standard deviations for the replicate series.
Figure 7
Comparison of the expansion in proteins identified between 1D (400 ng protein) and 2D LC−MS/MS (2 μg) experiments on normal renal and RCC FFPE extracts using the NanoAcquity UPLC-HDMS quadrupole-oa Tof (Waters, U.K.).
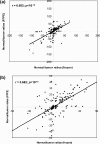
Figure 8
Comparison of normal/RCC tumor protein expression ratios (positive values = higher in tumor, negative = higher in normal tissue) derived from the label-free quantitative analyses of frozen versus FFPE tissue extracts (205 proteins in total). (a) All proteins plotted; (b) zoomed-in version of graph showing the region between 50:1 and 1:50.
Figure 9
Quantitative comparison of normal and tumor expression of several previously described proteins shown to be differentially expressed− in (a) fresh and (b) FFPE tissue extracts. Ratios calculated from deduced concentrations with reference to an internal standard in femtomoles/400 ng protein load using 1D-LC−MS are averaged from 3 injections and presented with their respective standard deviations.
Similar articles
- Equivalence of protein inventories obtained from formalin-fixed paraffin-embedded and frozen tissue in multidimensional liquid chromatography-tandem mass spectrometry shotgun proteomic analysis.
Sprung RW Jr, Brock JW, Tanksley JP, Li M, Washington MK, Slebos RJ, Liebler DC. Sprung RW Jr, et al. Mol Cell Proteomics. 2009 Aug;8(8):1988-98. doi: 10.1074/mcp.M800518-MCP200. Epub 2009 May 24. Mol Cell Proteomics. 2009. PMID: 19467989 Free PMC article. - Proteomic analysis of formalin-fixed paraffin-embedded renal tissue samples by label-free MS: assessment of overall technical variability and the impact of block age.
Craven RA, Cairns DA, Zougman A, Harnden P, Selby PJ, Banks RE. Craven RA, et al. Proteomics Clin Appl. 2013 Apr;7(3-4):273-82. doi: 10.1002/prca.201200065. Epub 2013 Mar 6. Proteomics Clin Appl. 2013. PMID: 23027403 - Quantitative Proteomic Analysis Using Formalin-Fixed, Paraffin-Embedded (FFPE) Human Cardiac Tissue.
Azimzadeh O, Atkinson MJ, Tapio S. Azimzadeh O, et al. Methods Mol Biol. 2021;2261:525-533. doi: 10.1007/978-1-0716-1186-9_33. Methods Mol Biol. 2021. PMID: 33421012 - Comparative evaluation of two methods for LC-MS/MS proteomic analysis of formalin fixed and paraffin embedded tissues.
Davalieva K, Kiprijanovska S, Dimovski A, Rosoklija G, Dwork AJ. Davalieva K, et al. J Proteomics. 2021 Mar 20;235:104117. doi: 10.1016/j.jprot.2021.104117. Epub 2021 Jan 14. J Proteomics. 2021. PMID: 33453434 Free PMC article. Review. - Complete solubilization of formalin-fixed, paraffin-embedded tissue may improve proteomic studies.
Shi SR, Taylor CR, Fowler CB, Mason JT. Shi SR, et al. Proteomics Clin Appl. 2013 Apr;7(3-4):264-72. doi: 10.1002/prca.201200031. Epub 2013 Mar 6. Proteomics Clin Appl. 2013. PMID: 23339100 Free PMC article. Review.
Cited by
- Proteomic and metabolic prediction of response to therapy in gastric cancer.
Aichler M, Luber B, Lordick F, Walch A. Aichler M, et al. World J Gastroenterol. 2014 Oct 14;20(38):13648-57. doi: 10.3748/wjg.v20.i38.13648. World J Gastroenterol. 2014. PMID: 25320503 Free PMC article. Review. - Reproducible proteomics sample preparation for single FFPE tissue slices using acid-labile surfactant and direct trypsinization.
Föll MC, Fahrner M, Oria VO, Kühs M, Biniossek ML, Werner M, Bronsert P, Schilling O. Föll MC, et al. Clin Proteomics. 2018 Mar 6;15:11. doi: 10.1186/s12014-018-9188-y. eCollection 2018. Clin Proteomics. 2018. PMID: 29527141 Free PMC article. - Tissue proteomics using chemical immobilization and mass spectrometry.
Shah P, Zhang B, Choi C, Yang S, Zhou J, Harlan R, Tian Y, Zhang Z, Chan DW, Zhang H. Shah P, et al. Anal Biochem. 2015 Jan 15;469:27-33. doi: 10.1016/j.ab.2014.09.017. Epub 2014 Oct 2. Anal Biochem. 2015. PMID: 25283129 Free PMC article. - Laser Capture Microdissection of Pancreatic Acinar Cells to Identify Proteomic Alterations in a Murine Model of Caerulein-Induced Pancreatitis.
Shapiro JP, Komar HM, Hancioglu B, Yu L, Jin M, Ogata Y, Hart PA, Cruz-Monserrate Z, Lesinski GB, Conwell DL. Shapiro JP, et al. Clin Transl Gastroenterol. 2017 Apr 13;8(4):e89. doi: 10.1038/ctg.2017.15. Clin Transl Gastroenterol. 2017. PMID: 28406494 Free PMC article. - Fast and Simple Protocols for Mass Spectrometry-Based Proteomics of Small Fresh Frozen Uterine Tissue Sections.
Dapic I, Uwugiaren N, Jansen PJ, Corthals GL. Dapic I, et al. Anal Chem. 2017 Oct 17;89(20):10769-10775. doi: 10.1021/acs.analchem.7b01937. Epub 2017 Oct 6. Anal Chem. 2017. PMID: 28910098 Free PMC article.
References
- Rifai N.; Gillette M. A.; Carr S. A. Protein biomarker discovery and validation: the long and uncertain path to clinical utility. Nat. Biotechnol. 2006, 24 (8), 971–983. - PubMed
- Bagnato C.; Thumar J.; Mayya V.; Hwang S. I.; Zebroski H.; Claffey K. P.; Haudenschild C.; Eng J. K.; Lundgren D. H.; Han D. K. Proteomics analysis of human coronary atherosclerotic plaque - A feasibility study of direct tissue proteomics by liquid chromatography and tandem mass spectrometry. Mol. Cell. Proteomics 2007, 6 (6), 1088–1102. - PubMed
- Hwang S. I.; Thumar J.; Lundgren D. H.; Rezaul K.; Mayya V.; Wu L.; Eng J.; Wright M. E.; Han D. K. Direct cancer tissue proteomics: a method to identify candidate cancer biomarkers from formalin-fixed paraffin-embedded archival tissues. Oncogene 2007, 26 (1), 65–76. - PubMed
- Palmer-Toy D. E.; et al. Efficient method for the proteomic analysis of fixed and embedded tissues. J. Proteome Res. 2005, 4 (6), 2404–2411. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- G0802416/MRC_/Medical Research Council/United Kingdom
- G0802416(89862)/MRC_/Medical Research Council/United Kingdom
- CRUK_/Cancer Research UK/United Kingdom
- DH_/Department of Health/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources