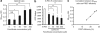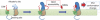Substrate docking to γ-secretase allows access of γ-secretase modulators to an allosteric site - PubMed (original) (raw)
Substrate docking to γ-secretase allows access of γ-secretase modulators to an allosteric site
Kengo Uemura et al. Nat Commun. 2010.
Free PMC article
Abstract
γ-Secretase generates the peptides of Alzheimer's disease, Aβ(40) and Aβ(42), by cleaving the amyloid precursor protein within its transmembrane domain. γ-Secretase also cleaves numerous other substrates, raising concerns about γ-secretase inhibitor off-target effects. Another important class of drugs, γ-secretase modulators, alter the cleavage site of γ-secretase on amyloid precursor protein, changing the Aβ(42)/Aβ(40) ratio, and are thus a promising therapeutic approach for Alzheimer's disease. However, the target for γ-secretase modulators is uncertain, with some data suggesting that they function on γ-secretase, whereas others support their binding to the amyloid precursor. In this paper we address this controversy by using a fluorescence resonance energy transfer-based assay to examine whether γ-secretase modulators alter Presenilin-1/γ-secretase conformation in intact cells in the absence of its natural substrates such as amyloid precursor protein and Notch. We report that the γ-secretase allosteric site is located within the γ-secretase complex, but substrate docking is needed for γ-secretase modulators to access this site.
Figures
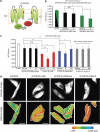
Figure 1. Substrate-dependent and -independent changes of PS1 conformation.
(a) Schematic structure of the G-PS1-R probe. GFP is fused to the PS1 N terminus (NT), and RFP is inserted into the large cytoplasmic loop between transmembrane domains 6 and 7. The red dots indicate D257 and D385 catalytic site aspartates. (b) FLIM analysis of the PS1 conformation in APP/APLP2 dKO cells (black bars) and in APP/APLP dKO cells reconstituted by stable expression of APP695 (APPdKO+695, green bars). GFP-PS1 (G-PS1)-transfected cells were used as a negative control to establish GFP lifetime in the absence of an acceptor fluorophore, which was comparable in APP/APLP2 dKO and APPdKO+695 cells (black and green bars, respectively). The GFP lifetime in G-PS1-R-transfected cells was compared in APP/APLP dKO and APP/APLP dKO+695 cells treated with vehicle or fenofibrate (ff) in three independent experiments (mean±s.d.; *P<0.001; NS, not significant; Fisher's PSLD, ANOVA; _n_=15–18 cells per condition were examined). (c) PS1 conformation was monitored in APP/APLP2 dKO cells co-transfected with G-PS1-R probe and either empty vector (black bars), C99 (red bars), NotchΔEC (blue bars) or CLAC-P (grey bars) constructs. The cells were treated with vehicle control, 100 μM fenofibrate (ff) or 400 μM ibuprofen (ibu) for 24 h. The graph shows mean±s.d. lifetime in psec; *P<0.05, **P<0.001; NS, not significant; ANOVA, _n_=3–5 independent experiments. On an average, 12–33 (vehicle), 17–20 (ff) and 12–18 (ibu) cells were examined. (d) The intensity images show GFP fluorescence reflecting the expression pattern of G-PS1-R probe. Pseudo-coloured FLIM images show subcellular distribution of the GFP lifetimes, with red pixels representing shorter lifetime (closer GFP-PS1 NT and RFP-PS1-loop proximity). Expression of the C99 substrate significantly increases red pixels, especially at the cell periphery. Ibuprofen reduces and fenofibrate increases the amount of red pixels, indicating 'opening' and 'closing' of the G-PS1-R conformation, respectively. The cell profiles are shown by tracing. A colourimetric scale bar shows colour-coded fluorescence lifetime in picoseconds.
Figure 2. Changes in G-PS1-R conformation correlate with changes in the Aβ42/40 ratio.
CHO cells stably expressing APP (7W cells) were transiently transfected with G-PS1-R probe and treated with the designated concentration of fenofibrate for 24 h. Conditioned media were subjected to Aβ ELISA (a), and the cells were used for the FLIM analysis of GFP lifetimes (G-PS1-R conformation, b). Fenofibrate treatment dose-dependently increased the Aβ42/40 ratio (a), and shortened GFP donor lifetime (b). The graph shows mean±s.d.; *P<0.05, **P<0.001; NS, not significant; ANOVA, _n_=3 independent experiments. (c) FRET efficiency was calculated using the following equation: E%=100*(_t_1−_t_2)/_t_1, where _t_1 is G-PS1 lifetime (FRET negative control, no acceptor) and _t_2 is a second, shorter GFP lifetime in G-PS1-R-transfected cells. E% is plotted against the Aβ42/40 ratio. The correlation coefficient=0.99678648 (regression line: _y_=0.023_x_−0.2127).
Figure 3. FLIM analysis of the changes in GFP-PS1-RFP conformation in APP/APLP2 dKO cells.
(a) Schematic representation of the helical peptide, HP and GSM binding site on APP C99. (b) GSMs affect PS1 conformation (NT-loop proximity) in the presence of HP. APP/APLP dKO cells transfected with G-PS1-R probe were treated with either HP alone or HP together with fenofibrate (ff) or ibuprofen (ibu). The graph shows mean±s.d. lifetime in picoseconds; *P<0.05, **P<0.001; ANOVA, _n_=3 independent experiments. On an average, 11–20 cells per condition were examined. (c) APP/APLP dKO cells were transfected with either wild-type G-PS1-R probe or G-PS1-R with FAD L166P mutation, and PS1 NT (GFP) to PS1-loop (RFP) proximity was analysed by FLIM (mean±s.d.; *P<0.05, ANOVA, _n_=3 independent experiments). On an average, 18–21 cells per condition were examined. (d) FLIM analysis of PS1 conformation in APP/APLP dKO cells co-transfected with G-PS1-R probe, together with either wild-type Pen-2 or N-terminally modified Flag-Pen-2 (mean±s.d.; *P<0.05, ANOVA, _n_=3 independent experiments). On an average, 14–16 cells per condition were examined.
Figure 4. Two-step model of the GSM action.
The allosteric site responsible for PS1 conformational change and located within the γ-secretase complex is hidden in the absence of APP C99/NotchΔEC substrate. On binding of the substrate to the docking site, the allosteric site is 'activated' and becomes accessible for GSMs. Subsequent action of GSMs on the γ-secretase allosteric site leads to 'opening' or 'closing' of PS1 conformation.
Similar articles
- γ-Secretase modulators exhibit selectivity for modulation of APP cleavage but inverse γ-secretase modulators do not.
Lessard CB, Rodriguez E, Ladd TB, Minter LM, Osborne BA, Miele L, Golde TE, Ran Y. Lessard CB, et al. Alzheimers Res Ther. 2020 May 19;12(1):61. doi: 10.1186/s13195-020-00622-5. Alzheimers Res Ther. 2020. PMID: 32430033 Free PMC article. - Allosteric modulation of PS1/gamma-secretase conformation correlates with amyloid beta(42/40) ratio.
Uemura K, Lill CM, Li X, Peters JA, Ivanov A, Fan Z, DeStrooper B, Bacskai BJ, Hyman BT, Berezovska O. Uemura K, et al. PLoS One. 2009 Nov 18;4(11):e7893. doi: 10.1371/journal.pone.0007893. PLoS One. 2009. PMID: 19924286 Free PMC article. - Inhibition of gamma-secretase as a therapeutic intervention for Alzheimer's disease: prospects, limitations and strategies.
Evin G, Sernee MF, Masters CL. Evin G, et al. CNS Drugs. 2006;20(5):351-72. doi: 10.2165/00023210-200620050-00002. CNS Drugs. 2006. PMID: 16696577 Review. - gamma-Secretase modulators.
Wolfe MS. Wolfe MS. Curr Alzheimer Res. 2007 Dec;4(5):571-3. doi: 10.2174/156720507783018299. Curr Alzheimer Res. 2007. PMID: 18220525 Review. - Quantification of gamma-secretase modulation differentiates inhibitor compound selectivity between two substrates Notch and amyloid precursor protein.
Yang T, Arslanova D, Gu Y, Augelli-Szafran C, Xia W. Yang T, et al. Mol Brain. 2008 Nov 4;1:15. doi: 10.1186/1756-6606-1-15. Mol Brain. 2008. PMID: 18983676 Free PMC article.
Cited by
- The brain-tumor related protein podoplanin regulates synaptic plasticity and hippocampus-dependent learning and memory.
Cicvaric A, Yang J, Krieger S, Khan D, Kim EJ, Dominguez-Rodriguez M, Cabatic M, Molz B, Acevedo Aguilar JP, Milicevic R, Smani T, Breuss JM, Kerjaschki D, Pollak DD, Uhrin P, Monje FJ. Cicvaric A, et al. Ann Med. 2016 Dec;48(8):652-668. doi: 10.1080/07853890.2016.1219455. Epub 2016 Aug 25. Ann Med. 2016. PMID: 27558977 Free PMC article. - Enzyme-substrate interface targeting by imidazole-based γ-secretase modulators activates γ-secretase and stabilizes its interaction with APP.
Petit D, Hitzenberger M, Koch M, Lismont S, Zoltowska KM, Enzlein T, Hopf C, Zacharias M, Chávez-Gutiérrez L. Petit D, et al. EMBO J. 2022 Nov 2;41(21):e111084. doi: 10.15252/embj.2022111084. Epub 2022 Sep 19. EMBO J. 2022. PMID: 36121025 Free PMC article. - Fibronectin type III domain-containing protein 5 interacts with APP and decreases amyloid β production in Alzheimer's disease.
Noda Y, Kuzuya A, Tanigawa K, Araki M, Kawai R, Ma B, Sasakura Y, Maesako M, Tashiro Y, Miyamoto M, Uemura K, Okuno Y, Kinoshita A. Noda Y, et al. Mol Brain. 2018 Oct 24;11(1):61. doi: 10.1186/s13041-018-0401-8. Mol Brain. 2018. PMID: 30355327 Free PMC article. - γ-Secretase modulator (GSM) photoaffinity probes reveal distinct allosteric binding sites on presenilin.
Pozdnyakov N, Murrey HE, Crump CJ, Pettersson M, Ballard TE, Am Ende CW, Ahn K, Li YM, Bales KR, Johnson DS. Pozdnyakov N, et al. J Biol Chem. 2013 Apr 5;288(14):9710-9720. doi: 10.1074/jbc.M112.398602. Epub 2013 Feb 8. J Biol Chem. 2013. PMID: 23396974 Free PMC article. - N-cadherin enhances APP dimerization at the extracellular domain and modulates Aβ production.
Asada-Utsugi M, Uemura K, Noda Y, Kuzuya A, Maesako M, Ando K, Kubota M, Watanabe K, Takahashi M, Kihara T, Shimohama S, Takahashi R, Berezovska O, Kinoshita A. Asada-Utsugi M, et al. J Neurochem. 2011 Oct;119(2):354-63. doi: 10.1111/j.1471-4159.2011.07364.x. Epub 2011 Sep 20. J Neurochem. 2011. PMID: 21699541 Free PMC article.
References
- De Strooper B. et al.. Deficiency of presenilin-1 inhibits the normal cleavage of amyloid precursor protein. Nature 391, 387–390 (1998). - PubMed
- Wolfe M. S. et al.. Two transmembrane aspartates in presenilin-1 required for presenilin endoproteolysis and gamma-secretase activity. Nature 398, 513–517 (1999). - PubMed
- Bitan G., Vollers S. S. & Teplow D. B. Elucidation of primary structure elements controlling early amyloid β-protein oligomerization. J. Biol. Chem. 278, 34882–34889 (2003). - PubMed
- Chen Y. R. & Glabe C. G. Distinct early folding and aggregation properties of Alzheimer amyloid-β peptides Aβ40 and Aβ42: stable trimer or tetramer formation by Aβ42. J. Biol. Chem. 281, 24414–24422 (2006). - PubMed
- Weggen S. et al.. Evidence that nonsteroidal anti-inflammatory drugs decrease amyloid β42 production by direct modulation of gamma-secretase activity. J. Biol. Chem. 278, 31831–31837 (2003). - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources