Readthrough of nonsense mutations in Rett syndrome: evaluation of novel aminoglycosides and generation of a new mouse model - PubMed (original) (raw)
Readthrough of nonsense mutations in Rett syndrome: evaluation of novel aminoglycosides and generation of a new mouse model
Cornelia Brendel et al. J Mol Med (Berl). 2011 Apr.
Erratum in
- J Mol Med (Berl). 2013 Jun;91(6):775
Abstract
Thirty-five percent of patients with Rett syndrome carry nonsense mutations in the MECP2 gene. We have recently shown in transfected HeLa cells that readthrough of nonsense mutations in the MECP2 gene can be achieved by treatment with gentamicin and geneticin. This study was performed to test if readthrough can also be achieved in cells endogenously expressing mutant MeCP2 and to evaluate potentially more effective readthrough compounds. A mouse model was generated carrying the R168X mutation in the MECP2 gene. Transfected HeLa cells expressing mutated MeCP2 fusion proteins and mouse ear fibroblasts isolated from the new mouse model were treated with gentamicin and the novel aminoglycosides NB30, NB54, and NB84. The localization of the readthrough product was tested by immunofluorescence. Readthrough of the R168X mutation in mouse ear fibroblasts using gentamicin was detected but at lower level than in HeLa cells. As expected, the readthrough product, full-length Mecp2 protein, was located in the nucleus. NB54 and NB84 induced readthrough more effectively than gentamicin, while NB30 was less effective. Readthrough of nonsense mutations can be achieved not only in transfected HeLa cells but also in fibroblasts of the newly generated Mecp2(R168X) mouse model. NB54 and NB84 were more effective than gentamicin and are therefore promising candidates for readthrough therapy in Rett syndrome patients.
Figures
Fig. 1
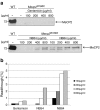
NB54- and NB84-mediated suppression of premature stop mutations. Western blot analysis of protein samples prepared from aminoglycoside-treated and untreated transiently transfected HeLa cells, using a monoclonal anti-FLAG antibody or a specific antibody detecting the C terminus of MeCP2. Thirty microgram of protein extract was loaded. Arrows denote the full-length FLAG-MeCP2 protein as well as the truncated FLAG-MeCP2 isoforms. As a loading control, GAPDH was detected in all samples. The cells were treated for 24 h with 500 μg/ml of gentamicin (a), NB30 (a), NB54 (a–b), or NB84 (b). The effect of 24-h drug treatment was quantified by densitometric western blot analysis from at least three independent experiments (c)
Fig. 2
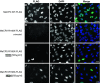
Nuclear localization of readthrough full-length MeCP2-FLAG fusion protein in drug-treated HeLa cells. Immunofluorescence of HeLa cells transfected with MeCP2-WT-FLAG- (a–c) compared to MeCP2-R168X-FLAG-expressing HeLa cells cultured under untreated (d–f) and drug-treated (g–i 500 μg/ml NB54; j–l 500 μg/ml NB84) conditions. Treatment was performed for 24 h. Localization of FLAG fusion proteins (left column) was visualized by using a monoclonal anti-FLAG antibody and immunofluorescence microscopy. Because the fusion protein was C-terminally FLAG-tagged, only full-length MeCP2 proteins were detected. The nuclei were counterstained with DAPI (middle column). Right column MeCP2-WT-FLAG expression as well as treated MeCP2-294-FLAG overlaps with DAPI staining, indicating a nuclear localization. Scale bars are 50 μm
Fig. 3
Gene targeting and Flp-mediated recombination at the Mecp2 locus. The diagram shows the wild-type Mecp2 genomic locus, the targeting vector, and the targeted locus before and after Flp-mediated recombination. The 5′ and 3′ regions of homology are shown in light gray. Small arrows indicate primers used to confirm gene targeting and Flp-mediated recombination, and to screen the genotype (a). Homologous recombination was verified by nested PCR of genomic DNA of the selected ES cell clones; primers 5/6 and 7/8 were as shown in a. The 1.9-kb band is from the mutated allele. The primer sets were designed not to amplify the wild-type Mecp2 gene. E5, E7, E32, and E38 are names of ES clones, while c and b indicate a plasmid DNA control demonstrating the mutant PCR fragment and blank. Primer set 9 and 10 were used for the genotyping PCR after FLP recombination (c). The 313-bp band represents the mutant allele (Hem hemizygous male, Het heterozygous female) containing the remaining FRT site, while the 265-bp band represents the wild-type allele (WT wild type). To confirm the presence of Mecp2 mRNA in the brain of two wild-type (WT (+/Y)) and two R168X (−/Y) mutant male mice, total RNA was extracted from brain of male wild-type and mutant mice and used for cDNA synthesis and RT-PCR (d). Western blot analysis (e) and immunofluorescent staining (f) demonstrated the absence of wild-type MeCP2 protein in nuclear extracts of hemizygous brains (Sup supernatant, Nuc nuclear extract) and in nuclei of mouse ear fibroblast using an antibody recognizing the N terminus of MeCP2. Note that no additional truncated protein was detected
Fig. 4
Drug-mediated re-expression of endogenous MeCP2. Western blot analysis of nuclear extracts prepared from aminoglycoside-treated and untreated mouse ear fibroblasts generated from wild-type and Mecp2 168 male mice: Immunoreactivity was detected by using a monoclonal anti-FLAG antibody and a specific antibody detecting the C terminus of MeCP2. Thirty microgram of protein extract was loaded into each lane. Arrows denote the full-length endogenous MeCP2 protein. The cells were treated for 4 days with 0–800 μg/ml of gentamicin, NB54, or NB84 (a). The effect of drug treatment was quantified by densitometric developed western blot analysis from at least three independent experiments (b)
Fig. 5
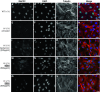
Nuclear localization of endogenous Mecp2 in drug-treated mouse fibroblasts. Immunofluorescent staining of mouse ear fibroblasts generated from wild-type and Mecp2 168 male mice. Cells were treated 4 days with 800 μg/ml gentamicin (i–l), NB54 (m–p), and NB84 (q–t), respectively. Localization of endogenous MeCP2 protein (first column) was visualized by using a specific MeCP2 antibody detecting the C-terminal part of the protein and immunofluorescent microscopy. The nuclei and the cytoskeleton of the cells were counterstained with DAPI (second column) and tubulin (third column). All three stainings are merged (last column). No MeCP2-specific staining was detected in untreated fibroblasts (e–h). Scale bars are 50 μm
Similar articles
- Ex vivo treatment with a novel synthetic aminoglycoside NB54 in primary fibroblasts from Rett syndrome patients suppresses MECP2 nonsense mutations.
Vecsler M, Ben Zeev B, Nudelman I, Anikster Y, Simon AJ, Amariglio N, Rechavi G, Baasov T, Gak E. Vecsler M, et al. PLoS One. 2011;6(6):e20733. doi: 10.1371/journal.pone.0020733. Epub 2011 Jun 13. PLoS One. 2011. PMID: 21695138 Free PMC article. - Evaluation of Novel Enhancer Compounds in Gentamicin-Mediated Readthrough of Nonsense Mutations in Rett Syndrome.
Wong KM, Wegener E, Baradaran-Heravi A, Huppke B, Gärtner J, Huppke P. Wong KM, et al. Int J Mol Sci. 2023 Jul 19;24(14):11665. doi: 10.3390/ijms241411665. Int J Mol Sci. 2023. PMID: 37511424 Free PMC article. - Systematic and quantitative analysis of stop codon readthrough in Rett syndrome nonsense mutations.
Lebeda D, Fierenz A, Werfel L, Rosin-Arbesfeld R, Hofhuis J, Thoms S. Lebeda D, et al. J Mol Med (Berl). 2024 May;102(5):641-653. doi: 10.1007/s00109-024-02436-6. Epub 2024 Mar 2. J Mol Med (Berl). 2024. PMID: 38430393 Free PMC article. - [Therapeutic readthrough strategy for suppression of nonsense mutations in duchenne muscular dystrophy].
Shiozuka M, Matsuda R. Shiozuka M, et al. Brain Nerve. 2011 Nov;63(11):1253-60. Brain Nerve. 2011. PMID: 22068478 Review. Japanese. - The MECP2 gene mutation screening in Rett syndrome patients from Croatia.
Matijević T, Knezević J, Barisić I, Resić B, Culić V, Pavelić J. Matijević T, et al. Ann N Y Acad Sci. 2006 Dec;1091:225-32. doi: 10.1196/annals.1378.069. Ann N Y Acad Sci. 2006. PMID: 17341617 Review.
Cited by
- Pharmacological read-through of R294X Mecp2 in a novel mouse model of Rett syndrome.
Merritt JK, Collins BE, Erickson KR, Dong H, Neul JL. Merritt JK, et al. Hum Mol Genet. 2020 Aug 29;29(15):2461-2470. doi: 10.1093/hmg/ddaa102. Hum Mol Genet. 2020. PMID: 32469049 Free PMC article. - Rett syndrome: insights into genetic, molecular and circuit mechanisms.
Ip JPK, Mellios N, Sur M. Ip JPK, et al. Nat Rev Neurosci. 2018 Jun;19(6):368-382. doi: 10.1038/s41583-018-0006-3. Nat Rev Neurosci. 2018. PMID: 29740174 Free PMC article. Review. - Respiratory phenotypes are distinctly affected in mice with common Rett syndrome mutations MeCP2 T158A and R168X.
Bissonnette JM, Schaevitz LR, Knopp SJ, Zhou Z. Bissonnette JM, et al. Neuroscience. 2014 May 16;267:166-76. doi: 10.1016/j.neuroscience.2014.02.043. Epub 2014 Mar 10. Neuroscience. 2014. PMID: 24626160 Free PMC article. - Defects in brainstem neurons associated with breathing and motor function in the Mecp2R168X/Y mouse model of Rett syndrome.
Johnson CM, Zhong W, Cui N, Wu Y, Xing H, Zhang S, Jiang C. Johnson CM, et al. Am J Physiol Cell Physiol. 2016 Dec 1;311(6):C895-C909. doi: 10.1152/ajpcell.00132.2016. Epub 2016 Sep 21. Am J Physiol Cell Physiol. 2016. PMID: 27653984 Free PMC article. - Early motor phenotype detection in a female mouse model of Rett syndrome is improved by cross-fostering.
Vogel Ciernia A, Pride MC, Durbin-Johnson B, Noronha A, Chang A, Yasui DH, Crawley JN, LaSalle JM. Vogel Ciernia A, et al. Hum Mol Genet. 2017 May 15;26(10):1839-1854. doi: 10.1093/hmg/ddx087. Hum Mol Genet. 2017. PMID: 28334953 Free PMC article.
References
- Rett A. On a unusual brain atrophy syndrome in hyperammonemia in childhood. Wien Med Wochenschr. 1946;116:723–726. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials