Visualization and statistical comparisons of microbial communities using R packages on Phylochip data - PubMed (original) (raw)
Visualization and statistical comparisons of microbial communities using R packages on Phylochip data
Susan Holmes et al. Pac Symp Biocomput. 2011.
Abstract
This article explains the statistical and computational methodology used to analyze species abundances collected using the LNBL Phylochip in a study of Irritable Bowel Syndrome (IBS) in rats. Some tools already available for the analysis of ordinary microarray data are useful in this type of statistical analysis. For instance in correcting for multiple testing we use Family Wise Error rate control and step-down tests (available in the multtest package). Once the most significant species are chosen we use the hypergeometric tests familiar for testing GO categories to test specific phyla and families. We provide examples of normalization, multivariate projections, batch effect detection and integration of phylogenetic covariation, as well as tree equalization and robustification methods.
Figures
Fig. 1
Tools were transposed from the standard microarray analyses
Fig. 2
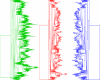
On the left the first plane of the PCA shows the first set of data with two batches and on the right the third set of arrays was added.
Fig. 3
On the left, we have the tree of all operational taxonomic units (otus) present on the Phylochip, we can observe that the distance to the root of many of the otus is variable, thus indicating a heterogeneous degree of resolution. The two trees on the right are filtered trees representing only the 400 most abundant species. The blue tree on the right was computed by using the collapsing algorithm presented in this section, we see that the long right clade at the bottom of the middle tree has disappeared.
Fig. 4
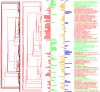
The left tree shows the complete tree on all species in black with the subtree of most abundant species in red, this subtree is the one plotted on the next panel. Values of abundance in CTL and IBS rats are plotted in the next two columns, the pink/blue scaled variables are the truncated rank differences between the two groups.
Fig. 5
Principal component analysis of top most abundant and variable species, we see the Mucosal location is the explanation for the first component, all the mucosal samples have negative loadings on this factor.
Fig. 6
Complete tree with the subtree of most variable among the consistently abundant species and the loadings on the first two principal components.
Fig. 7
The left tree shows the complete tree on all species in black with the subtree of set of species that show the most significantly differences between CTL and IBS in red in the second panel. Values of abundance in CTL and IBS rats are plotted in the next two columns, the next column shows the – log(pvalue), so the largest bars represent the most significantly different species.
Similar articles
- Application of phylogenetic microarrays to interrogation of human microbiota.
Paliy O, Agans R. Paliy O, et al. FEMS Microbiol Ecol. 2012 Jan;79(1):2-11. doi: 10.1111/j.1574-6941.2011.01222.x. Epub 2011 Nov 9. FEMS Microbiol Ecol. 2012. PMID: 22092522 Free PMC article. Review. - Gastrointestinal microbiome signatures of pediatric patients with irritable bowel syndrome.
Saulnier DM, Riehle K, Mistretta TA, Diaz MA, Mandal D, Raza S, Weidler EM, Qin X, Coarfa C, Milosavljevic A, Petrosino JF, Highlander S, Gibbs R, Lynch SV, Shulman RJ, Versalovic J. Saulnier DM, et al. Gastroenterology. 2011 Nov;141(5):1782-91. doi: 10.1053/j.gastro.2011.06.072. Epub 2011 Jul 8. Gastroenterology. 2011. PMID: 21741921 Free PMC article. - PhyloChip microarray analysis reveals altered gastrointestinal microbial communities in a rat model of colonic hypersensitivity.
Nelson TA, Holmes S, Alekseyenko AV, Shenoy M, Desantis T, Wu CH, Andersen GL, Winston J, Sonnenburg J, Pasricha PJ, Spormann A. Nelson TA, et al. Neurogastroenterol Motil. 2011 Feb;23(2):169-77, e41-2. doi: 10.1111/j.1365-2982.2010.01637.x. Epub 2010 Dec 3. Neurogastroenterol Motil. 2011. PMID: 21129126 Free PMC article. - A framework for analysis of metagenomic sequencing data.
Murat Eren A, Ferris MJ, Taylor CM. Murat Eren A, et al. Pac Symp Biocomput. 2011:131-41. doi: 10.1142/9789814335058_0015. Pac Symp Biocomput. 2011. PMID: 21121041 - Bioinformatics analysis of microarray data.
Zhang Y, Szustakowski J, Schinke M. Zhang Y, et al. Methods Mol Biol. 2009;573:259-84. doi: 10.1007/978-1-60761-247-6_15. Methods Mol Biol. 2009. PMID: 19763933 Review.
Cited by
- Application of phylogenetic microarrays to interrogation of human microbiota.
Paliy O, Agans R. Paliy O, et al. FEMS Microbiol Ecol. 2012 Jan;79(1):2-11. doi: 10.1111/j.1574-6941.2011.01222.x. Epub 2011 Nov 9. FEMS Microbiol Ecol. 2012. PMID: 22092522 Free PMC article. Review. - Prebiotic effects of diet supplemented with the cultivated red seaweed Chondrus crispus or with fructo-oligo-saccharide on host immunity, colonic microbiota and gut microbial metabolites.
Liu J, Kandasamy S, Zhang J, Kirby CW, Karakach T, Hafting J, Critchley AT, Evans F, Prithiviraj B. Liu J, et al. BMC Complement Altern Med. 2015 Aug 14;15:279. doi: 10.1186/s12906-015-0802-5. BMC Complement Altern Med. 2015. PMID: 26271359 Free PMC article. - Phyloseq: a bioconductor package for handling and analysis of high-throughput phylogenetic sequence data.
McMurdie PJ, Holmes S. McMurdie PJ, et al. Pac Symp Biocomput. 2012:235-46. Pac Symp Biocomput. 2012. PMID: 22174279 Free PMC article. - Machine learning applied to transcriptomic data to identify genes associated with feed efficiency in pigs.
Piles M, Fernandez-Lozano C, Velasco-Galilea M, González-Rodríguez O, Sánchez JP, Torrallardona D, Ballester M, Quintanilla R. Piles M, et al. Genet Sel Evol. 2019 Mar 13;51(1):10. doi: 10.1186/s12711-019-0453-y. Genet Sel Evol. 2019. PMID: 30866799 Free PMC article. - Establishing microbial composition measurement standards with reference frames.
Morton JT, Marotz C, Washburne A, Silverman J, Zaramela LS, Edlund A, Zengler K, Knight R. Morton JT, et al. Nat Commun. 2019 Jun 20;10(1):2719. doi: 10.1038/s41467-019-10656-5. Nat Commun. 2019. PMID: 31222023 Free PMC article.
References
- King T, Elia M, Hunter J. The Lancet. 1998 Jan; - PubMed
- Malinen E, Rinttilä T, Kajander K, Mättö J. The American journal of Gastroenterology. 2005 Jan;100:373. - PubMed
- Kassinen A, Krogius-Kurikka L, Mäkivuokko H. Gastroenterology. 2007 Jan; - PubMed