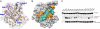Structure-function analyses point to a polynucleotide-accommodating groove essential for APOBEC3A restriction activities - PubMed (original) (raw)
. 2011 Feb;85(4):1765-76.
doi: 10.1128/JVI.01651-10. Epub 2010 Dec 1.
Iñigo Narvaiza, Alessandro Bertero, Shyam Peddi, Ute F Röhrig, Millán Ortiz, Vincent Zoete, Nataly Castro-Díaz, Priscilla Turelli, Amalio Telenti, Olivier Michielin, Matthew D Weitzman, Didier Trono
Affiliations
- PMID: 21123384
- PMCID: PMC3028873
- DOI: 10.1128/JVI.01651-10
Structure-function analyses point to a polynucleotide-accommodating groove essential for APOBEC3A restriction activities
Yannick Bulliard et al. J Virol. 2011 Feb.
Abstract
Members of the human APOBEC3 family of editing enzymes can inhibit various mobile genetic elements. APOBEC3A (A3A) can block the retrotransposon LINE-1 and the parvovirus adeno-associated virus type 2 (AAV-2) but does not inhibit retroviruses. In contrast, APOBEC3G (A3G) can block retroviruses but has only limited effects on AAV-2 or LINE-1. What dictates this differential target specificity remains largely undefined. Here, we modeled the structure of A3A based on its homology with the C-terminal domain of A3G and further compared the sequence of human A3A to those of 11 nonhuman primate orthologues. We then used these data to perform a mutational analysis of A3A, examining its ability to restrict LINE-1, AAV-2, and foreign plasmid DNA and to edit a single-stranded DNA substrate. The results revealed an essential functional role for the predicted single-stranded DNA-docking groove located around the A3A catalytic site. Within this region, amino acid differences between A3A and A3G are predicted to affect the shape of the polynucleotide-binding groove. Correspondingly, transferring some of these A3A residues to A3G endows the latter protein with the ability to block LINE-1 and AAV-2. These results suggest that the target specificity of APOBEC3 family members is partly defined by structural features influencing their interaction with polynucleotide substrates.
Figures
FIG. 1.
Alignment of human A3A and A3G C terminus. Color bars below the alignment reflect predicted secondary-structure elements (α-helices in orange and β-strands in green), while loop domains are simply numbered (in blue) from 1 to 7. Red stars indicate residues selected for site-directed mutagenesis on human A3A; black arrowheads indicate zinc-coordinating residues.
FIG. 2.
Homology modeling of human A3A. (A) Comparison of the ANOLEA scores of the A3A model (red) and the A3G X-ray structure 3IR2 (black). (B) Superposition of the A3A model (brown) and the A3G C-terminal X-ray structure (light blue). The zinc atom (yellow) and residues required for zinc coordination are illustrated. (C) Alignment of human A3A together with 11 novel primate orthologues, with mutated residues analyzed for restriction activity marked with an asterisk. Color bars below the alignment reflect predicted secondary-structure elements. Missing codons due to amplification failure are indicated with an X. Dark blue, >80% identity; blue, >60%; light blue, >40%. Alignment was performed using MUSCLE. Magenta stars indicate residues selected for site-directed mutagenesis on human A3A; black arrowheads indicate zinc-coordinating residues. AGM, African green monkey. (D) Residues selected for site-directed mutagenesis (magenta) mapped on the A3A homology model. The four loops flanking the catalytic site are shown in blue.
FIG. 3.
LINE-1, AAV, and foreign DNA restriction by wild-type and mutant A3A. (A) Levels of expression of transiently expressed A3A at three different doses for the wild type, while mutants were tested at the intermediate dose of plasmid DNA (0.2 μg), as analyzed by Western blotting using HA- and tubulin-specific antibodies. Two mutants with a steady-state level of expression that was reduced by 3-fold or more compared to that of the wild type are highlighted in boldface (top blot, far right). (B) Top, exon junction-spanning primers (black half arrows) selectively amplify the spliced neomycin resistance cassette obtained after LINE-1-mediated reverse transcription. Neo, _neo_-carrying plasmid; neo+intron, LINE-1 reporter plasmid; SD, splice donor; SA, splice acceptor. Bottom, experimental setup of a typical LINE-1 retrotransposition assay. Tfx, transfection. (C) LINE-1 retrotransposition efficiency as measured by semiquantitative PCR analysis of reverse-transcribed cDNA (bottom) or by the selection of neomycin-resistant colonies (top) in the absence or presence of the wild type or stably expressed yet functionally defective A3A mutants. Means and standard errors are representatives of independent duplicates. The production of rAAVLuc (D) and expression of GFP (E) in the presence or absence of the A3A derivatives also are shown. The amount of A3A-expressing plasmids used is indicated. In panel D, results from three independent experiments are presented as the averages of the relative values compared to those of mock transfections with pcDNA3.1(+).
FIG. 4.
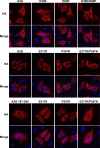
Subcellular localization of wild-type A3A and functionally defective mutants. APOBEC3 protein localization was determined by the indirect immunofluorescence of HA-tagged APOBEC3 proteins (HA) expressed in U2OS cells. Cells were costained with DAPI to visualize nuclei (Merge). U2OS cells were grown on glass coverslips and transfected with 0.8 μg APOBEC3 expression vector using Lipofectamine 2000 (Invitrogen). After 24 h, cells were fixed with 3% paraformaldehyde and extracted with 0.5% Triton X-100 in PBS. Cells were incubated with anti-HA MAb 16b12 (1:2,000) (Covance) and a 1:2,000 dilution of goat anti-mouse-conjugated Alexa Fluor 568 (Invitrogen) and DAPI (Sigma Aldrich). The coverslips were mounted in Fluoromount G (Southern Biotech), and images were acquired using a Leica TCS SP2 confocal microscope.
FIG. 5.
Inhibition of LINE-1 retrotransposition by A3A partially correlates with interference with plasmid DNA. (A) LINE-1 transcription and reverse transcription products on day 2 posttransfection, in the presence or absence of A3A and nucleoside reverse transcriptase inhibitors (NRTI). AZT, 40 μM; 3TC, 20 μM. Actin DNA served as a loading control and water as a negative control (H2O). Samples run in the absence of reverse transcriptase failed to yield any signal (not illustrated). (B) Determination of the number of neomycin- or hygromycin-resistant colonies in the presence of the LINE-1 reporter or a _neo_-carrying plasmid, as well as cell viability in the absence of G418. The transfection conditions among the four assays were essentially the same, except that the LINE-1-expressing plasmid in the first assay was replaced by a _neo_-carrying plasmid in the latter two assays. Black columns correspond to the A3A plasmid dose used in most functional assays to test A3A mutants.
FIG. 6.
Molecular docking of a single-stranded DNA template and cytidine deaminase assay. (A) Residues important (orange) or dispensable (magenta) for A3A restriction activity mapped on the structural model. Yellow, zinc-coordinating residues. (B) Constrained molecular docking of a 5′-TT
C
A oligonucleotide (cyan) onto the A3A model using EADocks. (C) Deaminase activity of A3A and mutant proteins generated by _in vitro_-coupled transcription-translation (IVT) was analyzed in UDG-dependent deaminase assays. Arrows indicate substrate deoxyoligonucleotide and cleaved deaminated product. Bottom panel, Western blot of one-fifth of the IVT reaction mixtures using HA-specific antibody.
FIG. 7.
Structure and primate sequence comparisons reveal a distinct DNA-binding groove on human A3A and the A3G C terminus. (A) Space-filling visualization showing the surface formed by the residues important for editing A3A or that are important for editing and are present on loops 1, 3, 5, and 6 for the A3G C terminus (orange), placed on the homology model or the X-ray structure. The catalytic-site pocket is highlighted in yellow. (B) Alignment of the 12 primate A3A orthologues with their A3G C-terminal counterparts, focusing on the four loops represented in Fig. 1D and on helix 6, the C-terminal-most helix of A3A. Orange and magenta stars indicate residues with or without functional importance, respectively; black frames indicate K30 and K60; black arrowheads indicate zinc-coordinating residues. Missing codons due to amplification failure are indicated with an X. Note that human A3G editing-defective mutants on helix 6 (at positions 364, 375, 378, and 379 [13]) are not depicted. (C) LINE-1 and AAV-2 replication efficiency and GFP expression in the presence of derivatives of the A3G C-terminal domain (residues 197 to 384). Means and standard errors are representative of the results for independent duplicates from three (LINE), two (AAV), or one experiment (GFP). The bottom shows expression levels of the A3G C-terminal derivatives and A3A, as evaluated by Western blot analysis. (D) LINE-1 retrotransposition efficiency, depicted as described for panel C, except that full-length A3G (residues 1 to 384; left) or A3A (right) was used.
FIG. 8.
Subcellular localization of wild-type A3A, full-length A3G, and the C terminus of A3G and the indicated mutants.
Similar articles
- Deaminase-independent inhibition of parvoviruses by the APOBEC3A cytidine deaminase.
Narvaiza I, Linfesty DC, Greener BN, Hakata Y, Pintel DJ, Logue E, Landau NR, Weitzman MD. Narvaiza I, et al. PLoS Pathog. 2009 May;5(5):e1000439. doi: 10.1371/journal.ppat.1000439. Epub 2009 May 22. PLoS Pathog. 2009. PMID: 19461882 Free PMC article. - A DNA sequence recognition loop on APOBEC3A controls substrate specificity.
Logue EC, Bloch N, Dhuey E, Zhang R, Cao P, Herate C, Chauveau L, Hubbard SR, Landau NR. Logue EC, et al. PLoS One. 2014 May 14;9(5):e97062. doi: 10.1371/journal.pone.0097062. eCollection 2014. PLoS One. 2014. PMID: 24827831 Free PMC article. - Structural determinants of human APOBEC3A enzymatic and nucleic acid binding properties.
Mitra M, Hercík K, Byeon IJ, Ahn J, Hill S, Hinchee-Rodriguez K, Singer D, Byeon CH, Charlton LM, Nam G, Heidecker G, Gronenborn AM, Levin JG. Mitra M, et al. Nucleic Acids Res. 2014 Jan;42(2):1095-110. doi: 10.1093/nar/gkt945. Epub 2013 Oct 24. Nucleic Acids Res. 2014. PMID: 24163103 Free PMC article. - Role of the single deaminase domain APOBEC3A in virus restriction, retrotransposition, DNA damage and cancer.
Wang Y, Schmitt K, Guo K, Santiago ML, Stephens EB. Wang Y, et al. J Gen Virol. 2016 Jan;97(1):1-17. doi: 10.1099/jgv.0.000320. Epub 2015 Oct 20. J Gen Virol. 2016. PMID: 26489798 Free PMC article. Review. - Regulation, functional impact, and therapeutic targeting of APOBEC3A in cancer.
Kawale AS, Zou L. Kawale AS, et al. DNA Repair (Amst). 2024 Sep;141:103734. doi: 10.1016/j.dnarep.2024.103734. Epub 2024 Jul 20. DNA Repair (Amst). 2024. PMID: 39047499 Review.
Cited by
- Binding of RNA by APOBEC3G controls deamination-independent restriction of retroviruses.
Bélanger K, Savoie M, Rosales Gerpe MC, Couture JF, Langlois MA. Bélanger K, et al. Nucleic Acids Res. 2013 Aug;41(15):7438-52. doi: 10.1093/nar/gkt527. Epub 2013 Jun 12. Nucleic Acids Res. 2013. PMID: 23761443 Free PMC article. - Mechanisms for targeted, purposeful mutation revealed in an APOBEC-DNA complex.
Schutsky EK, Hostetler ZM, Kohli RM. Schutsky EK, et al. Nat Struct Mol Biol. 2017 Feb 6;24(2):97-98. doi: 10.1038/nsmb.3373. Nat Struct Mol Biol. 2017. PMID: 28169999 No abstract available. - APOBEC3A associates with human papillomavirus genome integration in oropharyngeal cancers.
Kondo S, Wakae K, Wakisaka N, Nakanishi Y, Ishikawa K, Komori T, Moriyama-Kita M, Endo K, Murono S, Wang Z, Kitamura K, Nishiyama T, Yamaguchi K, Shigenobu S, Muramatsu M, Yoshizaki T. Kondo S, et al. Oncogene. 2017 Mar 23;36(12):1687-1697. doi: 10.1038/onc.2016.335. Epub 2016 Oct 3. Oncogene. 2017. PMID: 27694899 - An overview of the functions and mechanisms of APOBEC3A in tumorigenesis.
Yang Y, Liu N, Gong L. Yang Y, et al. Acta Pharm Sin B. 2024 Nov;14(11):4637-4648. doi: 10.1016/j.apsb.2024.08.020. Epub 2024 Aug 27. Acta Pharm Sin B. 2024. PMID: 39664421 Free PMC article. Review. - Functions and consequences of AID/APOBEC-mediated DNA and RNA deamination.
Pecori R, Di Giorgio S, Paulo Lorenzo J, Nina Papavasiliou F. Pecori R, et al. Nat Rev Genet. 2022 Aug;23(8):505-518. doi: 10.1038/s41576-022-00459-8. Epub 2022 Mar 7. Nat Rev Genet. 2022. PMID: 35256818 Free PMC article. Review.
References
- Aguiar, R. S., N. Lovsin, A. Tanuri, and B. M. Peterlin. 2008. Vpr.A3A chimera inhibits HIV replication. J. Biol. Chem. 283:2518-2525. - PubMed
- Bishop, K. N., et al. 2004. Cytidine deamination of retroviral DNA by diverse APOBEC proteins. Curr. Biol. 14:1392-1396. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases