Dlk1 is necessary for proper skeletal muscle development and regeneration - PubMed (original) (raw)
Dlk1 is necessary for proper skeletal muscle development and regeneration
Jolena N Waddell et al. PLoS One. 2010.
Abstract
Delta-like 1homolog (Dlk1) is an imprinted gene encoding a transmembrane protein whose increased expression has been associated with muscle hypertrophy in animal models. However, the mechanisms by which Dlk1 regulates skeletal muscle plasticity remain unknown. Here we combine conditional gene knockout and over-expression analyses to investigate the role of Dlk1 in mouse muscle development, regeneration and myogenic stem cells (satellite cells). Genetic ablation of Dlk1 in the myogenic lineage resulted in reduced body weight and skeletal muscle mass due to reductions in myofiber numbers and myosin heavy chain IIB gene expression. In addition, muscle-specific Dlk1 ablation led to postnatal growth retardation and impaired muscle regeneration, associated with augmented myogenic inhibitory signaling mediated by NF-κB and inflammatory cytokines. To examine the role of Dlk1 in satellite cells, we analyzed the proliferation, self-renewal and differentiation of satellite cells cultured on their native host myofibers. We showed that ablation of Dlk1 inhibits the expression of the myogenic regulatory transcription factor MyoD, and facilitated the self-renewal of activated satellite cells. Conversely, Dlk1 over-expression inhibited the proliferation and enhanced differentiation of cultured myoblasts. As Dlk1 is expressed at low levels in satellite cells but its expression rapidly increases upon myogenic differentiation in vitro and in regenerating muscles in vivo, our results suggest a model in which Dlk1 expressed by nascent or regenerating myofibers non-cell autonomously promotes the differentiation of their neighbor satellite cells and therefore leads to muscle hypertrophy.
Conflict of interest statement
Competing Interests: The authors have declared that no competing interests exist.
Figures
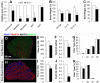
Figure 1. Myf5-Cre mediated mutation of paternal Dlk1(cKO) results in defects in muscle formation and growth.
Asterisks in all graphs denote p<0.05compared to wild-type (WT) controls. A: Quantitative PCR confirming muscle-specific Dlk1 knockout in cKO mice in whole muscle, myofibers, myoblasts and BAT. B: Relative body weight (BW) of WT (n = 17, 9, 4 for all age, 2–4 month and 10–12 month mice, respectively) and cKO (n = 18, 9, 5) littermates. C: Relative mass of BAT of WT (n = 3) and cKO (n = 3) mice. D–K: Muscle fiber composition in representative fast-twitch (EDL) and slow-twitch soleus (SOL) muscles revealed by MyHC isoform-specific antibodies and qPCR. D: MyHC isoform staining of representative EDL. Total myofiber number for EDL(E) and SOL (I) muscle in WT and cKO (n = 4 pairs). Myosin heavy chain (MyHC) IIB gene expression in EDL (F) and SOL (J;n = 3 pairs) muscles. Percent of each MyHC isoform by immunostaining in EDL (G) and SOL (K; n = 3 pairs).
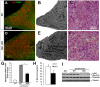
Figure 2. Muscle-specific Dlk1 cKO results in impaired muscle regeneration after injury.
A–F: Morphology of TA muscle 1 week after cardiotoxin injury in WT (A–C) and cKO mice (D–F). A, D: Fluorescent immunostaining of mouse IgG showing injured fibers in red and Dapi nuclei staining in green. B, E: Brightfield images showing increased fibrosis and scarification (Dark signal due to poor light penetration) in cKO mutant muscles. C, F: H–E staining showing poor organization and increased infiltration of non-myogenic cells in mutant muscles. G: Myogenin mRNA levels increase after injury, but cKO mice have an impaired myogenin response (n = 4). H: Total number of nascent fibers (n = 7 pairs) are decreased in cKO injured muscles. I: Levels of phosphorylated Akt are decreased in cKO muscle compared to WT control and injured muscles.
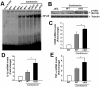
Figure 3. Muscle-specific Dlk1 cKO results in increased inflammation and NF-κB signaling after injury.
A–B: NF-κB pathway is up-regulated in cKO injured muscle as indicated by increase DNA binding by nuclear NF-κB (A) and phosphorylated Iκ-Bα (B). C: CD68 (macrophage marker) mRNA levels are increased in cKO injured muscles (n = 4). D–E: Pro-inflammatory cytokines IL-1β (D) and TNF-α (E) are up-regulated in cKO injured muscles (n = 4).
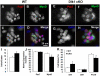
Figure 4. Dlk1 regulates satellite cell self-renewal and differentiation.
A–D: Pax7 and MyoD expression in a representative cluster of myoblasts on a wild-type EDL fiber after 3 days of culture. Pax7+/MyoD−, Pax7+/MyoD+ and Pax7−/MyoD+ cells represent self-renewing, proliferating and differentiating cells, respectively. E–H: A representative cluster of myoblasts on a cKO fiber cultured under identical culture conditions. I: The average number of cells per cluster. J: The percentage of cells expressing Pax7 and MyoD (note that most cells co-express Pax7 and MyoD; see K below). K: The proportion of cells at three different statuses: self-renewal (S–R; Pax7+/MyoD−), differentiation (Diff; Pax7−/MyoD+) and proliferation (Prolif; Pax7+/MyoD+). In I–K, n = 34 for WT, and n = 27 for cKO; * indicates p<0.05 compared to WT.
Figure 5. Dlk1 over-expression (OE) or recombinant Dlk1 protein inhibits proliferation but promotes differentiation of myoblasts.
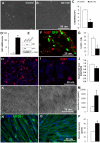
A–B: Dlk1 over-expression in C2C12 myoblasts. Cells were transfected with either GFP (Control; A) or Dlk1 OE (B) plasmids and cultured for 3 days. C: Cell numbers of Control and Dlk1 OE at day 3 after transfection. D–E: Relative Dlk1 mRNA levels in the Control and Dlk1 OE cells measured by quantitative Realtime PCR (n = 4). E: Protein levels of Dlk1, GFP and α-tubulin in GFP control transfected cells (GFP), Dlk1 over-expression cells (DLK1), and cells transfected with an unrelated negative control gene (TSG101). F: Primary myoblasts co-transfected with Dlk1 and GFP (4∶1) plasmids were cultured for 2 days and labeled with a cell proliferation marker Ki67 together with GFP staining. GFP signals (indicating Dlk1 positively transfected cells) exhibit little colocalized with Ki67 immunofluorescence. G: Percentage of cells displaying Ki67 staining in Control and Dlk1 OE cells (n = 3). H–J: Primary myoblasts transfected with an empty plasmid (H, control) or Dlk1 plasmid (I, OE) cultured for 3 days in growth medium and labeled with Ki67 (in red) and DAPI (in blue). The average Ki67 immunofluorescenc intensity was measured with Photoshop and normalized to DAPI intensity (J, n = 3 per treatment). K–M: Phase-contrast images of primary myoblasts differentiated for 48 hrs after transfected with either empty plasmids (K) or Dlk1 OE plasmids (L). M: Average pixel size of myotubes in Control and Dlk1 OE treatments as shown in H&I (n = 20 random tubes measured). N–O: Primary myoblasts grown on Matrigel plus vehicle control (N) and on Matrigel plus Dlk1 recombinant protein (O) after 3 days in differentiation medium. Green fluorescence (MF20) marks sarcomere myosin heavy chain and Blue is DAPI counterstaining for nuclei. P: The relative diameters of the resulting myotubes as shown in N & O were measured with Image J software. Control is the open bar (n = 66 myotubes) and Dlk1 recombinant protein treated cells are represented by the solid black bar (n = 66 myotubes). Asterisks in all bar graphs indicate p<0.05 compared to control groups by student t-test.
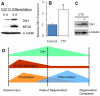
Figure 6. Dlk1 up-regulation correlates to myogenic differentiation and skeletal muscle regeneration after injury.
A: Representative Western-blot image showing that Dlk1 protein levels increase 5 days after serum-withdrawal induced differentiation of C2C12 myoblasts. B: The relative level of Dlk1 mRNA in the TA muscle revealed by quantitative Realtime PCR 5 days after cardiotoxin-induced regeneration (* indicates P<0.05; n = 18 for control TA; 6 for CTX treated TA). C: Dlk1 protein expression revealed by Western-blot in resting and regenerating TA muscles 5 days after cardiotoxin treatment. D: A model depicting how Dlk1 level varies during muscle quiescence and regeneration, which correlates to suppression of NK-κB activity and the onset of myoblast differentiation.
Similar articles
- CCAAT/enhancer binding protein β is required for satellite cell self-renewal.
Lala-Tabbert N, AlSudais H, Marchildon F, Fu D, Wiper-Bergeron N. Lala-Tabbert N, et al. Skelet Muscle. 2016 Dec 7;6(1):40. doi: 10.1186/s13395-016-0112-8. Skelet Muscle. 2016. PMID: 27923399 Free PMC article. - Xin, an actin binding protein, is expressed within muscle satellite cells and newly regenerated skeletal muscle fibers.
Hawke TJ, Atkinson DJ, Kanatous SB, Van der Ven PF, Goetsch SC, Garry DJ. Hawke TJ, et al. Am J Physiol Cell Physiol. 2007 Nov;293(5):C1636-44. doi: 10.1152/ajpcell.00124.2007. Epub 2007 Sep 13. Am J Physiol Cell Physiol. 2007. PMID: 17855775 - The transcriptional co-repressor TLE3 regulates myogenic differentiation by repressing the activity of the MyoD transcription factor.
Kokabu S, Nakatomi C, Matsubara T, Ono Y, Addison WN, Lowery JW, Urata M, Hudnall AM, Hitomi S, Nakatomi M, Sato T, Osawa K, Yoda T, Rosen V, Jimi E. Kokabu S, et al. J Biol Chem. 2017 Aug 4;292(31):12885-12894. doi: 10.1074/jbc.M116.774570. Epub 2017 Jun 12. J Biol Chem. 2017. PMID: 28607151 Free PMC article. - Skeletal muscle hypertrophy and regeneration: interplay between the myogenic regulatory factors (MRFs) and insulin-like growth factors (IGFs) pathways.
Zanou N, Gailly P. Zanou N, et al. Cell Mol Life Sci. 2013 Nov;70(21):4117-30. doi: 10.1007/s00018-013-1330-4. Epub 2013 Apr 4. Cell Mol Life Sci. 2013. PMID: 23552962 Free PMC article. Review. - Defining the transcriptional signature of skeletal muscle stem cells.
Yablonka-Reuveni Z, Day K, Vine A, Shefer G. Yablonka-Reuveni Z, et al. J Anim Sci. 2008 Apr;86(14 Suppl):E207-16. doi: 10.2527/jas.2007-0473. Epub 2007 Sep 18. J Anim Sci. 2008. PMID: 17878281 Free PMC article. Review.
Cited by
- Delta-like 1 homolog (dlk1): a marker for rhabdomyosarcomas implicated in skeletal muscle regeneration.
Jørgensen LH, Sellathurai J, Davis EE, Thedchanamoorthy T, Al-Bader RW, Jensen CH, Schrøder HD. Jørgensen LH, et al. PLoS One. 2013;8(4):e60692. doi: 10.1371/journal.pone.0060692. Epub 2013 Apr 5. PLoS One. 2013. PMID: 23577150 Free PMC article. - Proteome and transcriptome profiling of equine myofibrillar myopathy identifies diminished peroxiredoxin 6 and altered cysteine metabolic pathways.
Valberg SJ, Perumbakkam S, McKenzie EC, Finno CJ. Valberg SJ, et al. Physiol Genomics. 2018 Dec 1;50(12):1036-1050. doi: 10.1152/physiolgenomics.00044.2018. Epub 2018 Oct 5. Physiol Genomics. 2018. PMID: 30289745 Free PMC article. - Consequences of MEGF10 deficiency on myoblast function and Notch1 interactions.
Saha M, Mitsuhashi S, Jones MD, Manko K, Reddy HM, Bruels CC, Cho KA, Pacak CA, Draper I, Kang PB. Saha M, et al. Hum Mol Genet. 2017 Aug 1;26(15):2984-3000. doi: 10.1093/hmg/ddx189. Hum Mol Genet. 2017. PMID: 28498977 Free PMC article. - Inducible Deletion of Protein Kinase Map4k4 in Obese Mice Improves Insulin Sensitivity in Liver and Adipose Tissues.
Danai LV, Flach RJ, Virbasius JV, Menendez LG, Jung DY, Kim JH, Kim JK, Czech MP. Danai LV, et al. Mol Cell Biol. 2015 Jul;35(13):2356-65. doi: 10.1128/MCB.00150-15. Mol Cell Biol. 2015. PMID: 25918248 Free PMC article. - MEF2A regulates the Gtl2-Dio3 microRNA mega-cluster to modulate WNT signaling in skeletal muscle regeneration.
Snyder CM, Rice AL, Estrella NL, Held A, Kandarian SC, Naya FJ. Snyder CM, et al. Development. 2013 Jan 1;140(1):31-42. doi: 10.1242/dev.081851. Epub 2012 Nov 15. Development. 2013. PMID: 23154418 Free PMC article.
References
- Floridon C, Jensen CH, Thorsen P, Nielsen O, Sunde L, et al. Does Fetal antigen 1 (FA1) identify cells with regenerative, endocrine and neuroendocrine potentials? A study of FA1 in embryonic, fetal, and placental tissue and in maternal circulation. Differentiation. 2000;66:49–59. - PubMed
- Davis E, Jensen CH, Schroder HD, Farnir F, Shay-Hadfield T, et al. Ectopic Expression of DLK1 Protein in Skeletal Muscle of Padumnal Heterozygotes Causes the Callipyge Phenotype. Curr Biol. 2004;14:1858–1862. - PubMed
- White JD, Vuocolo T, McDonagh M, Grounds MD, Harper GS, et al. Analysis of the callipyge phenotype through skeletal muscle development; association of Dlk1 with muscle precursor cells. Differentiation. 2008;76:283–298. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 AR060652-01A1/AR/NIAMS NIH HHS/United States
- R01 AR060652/AR/NIAMS NIH HHS/United States
- HD042013/HD/NICHD NIH HHS/United States
- R01 HD042013/HD/NICHD NIH HHS/United States
- 1R01AR060652-01A1/AR/NIAMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials