Autophagy and metabolism - PubMed (original) (raw)
Review
Autophagy and metabolism
Joshua D Rabinowitz et al. Science. 2010.
Abstract
Autophagy is a process of self-cannibalization. Cells capture their own cytoplasm and organelles and consume them in lysosomes. The resulting breakdown products are inputs to cellular metabolism, through which they are used to generate energy and to build new proteins and membranes. Autophagy preserves the health of cells and tissues by replacing outdated and damaged cellular components with fresh ones. In starvation, it provides an internal source of nutrients for energy generation and, thus, survival. A powerful promoter of metabolic homeostasis at both the cellular and whole-animal level, autophagy prevents degenerative diseases. It does have a downside, however--cancer cells exploit it to survive in nutrient-poor tumors.
Figures
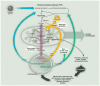
Fig. 1
Use of the products of autophagy. Multiple forms of stress activate autophagy (bottom right). Degradation of proteins, lipids, carbohydrates, and nucleic acids liberates amino acids, fatty acids, sugars, and nucleosides that are released into the cytoplasm for reutilization. Sugars (blue lines), including glucose released from glycogen granules by glycogenolysis or autophagy, are catabolized by glycolysis and the PPP to generate ATP, and pyruvate for subsequent TCA cycle metabolism. Nucleosides (green lines) are used for new nucleic acid synthesis and catabolized by the combined action of the PPP and glycolysis. Amino acids (purple lines) are used as building blocks for new protein synthesis, for ATP production by central carbon metabolism, and (in liver) as substrates for gluconeogenesis (Fig. 3). They also can be combined to yield citrate, which drives lipid synthesis and membrane biogenesis. Catabolism of amino acids yields ammonia, an activator of autophagy (dotted line). Fatty acids (yellow lines) from lipolysis or from autophagy of membranes or lipid droplets yield acetyl-CoA, which feeds the TCA cycle, supporting ATP production and citrate generation. OAA indicates oxaloacetate; α-KG, α-ketoglutarate; and ER, endoplasmic reticulum.
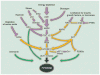
Fig. 2
Signaling pathways that regulate autophagy. Common nutrient, growth factor, hormone, and stress signals that regulate autophagy. Purple lines depict events that positively regulate autophagy. Yellow lines depict those that negatively regulate autophagy. Many pathways converge on the AMPK-mTORC1 axis. Green lines depict pathways that are mTOR-independent. Note that all input signals are framed as autophagy activators; thus, they include limitation for growth factors and nutrients. IKKβ, inhibitor of nuclear factor κB kinase β; PI3K, phosphatidylinositol-3 kinase; PTEN, phosphatase and tensin homolog; MAPK, mitogen-activated protein kinase; TSC1/2, tuberosclerosis complexes 1 and 2; and EF, elongation factor.
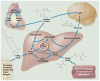
Fig. 3
Role of autophagy in adult mammalian starvation. Depicted pathways predominate after depletion of glycogen stores, typically ~12 hours into starvation. Autophagy in liver and heart (but not brain) generates fatty acids and amino acids, which are catabolized to yield energy. In the liver, this energy drives gluconeogenesis and ketogenesis. Amino acids are substrates for both ketogenesis and gluconeogenesis; acetyl-CoA from fatty acids is only for ketogenesis. As starvation continues, degradation of adipose and muscle play an increasing role in supplying substrates to the liver, which exports glucose and ketone bodies to feed the brain. The relative importance of ketone bodies increases in prolonged starvation. NADH, reduced form of nicotinamide adenine dinucleotide.
Similar articles
- Nutrient-sensing mTORC1: Integration of metabolic and autophagic signals.
Tan VP, Miyamoto S. Tan VP, et al. J Mol Cell Cardiol. 2016 Jun;95:31-41. doi: 10.1016/j.yjmcc.2016.01.005. Epub 2016 Jan 7. J Mol Cell Cardiol. 2016. PMID: 26773603 Free PMC article. Review. - Together we are stronger: fusion protects mitochondria from autophagosomal degradation.
Rambold AS, Kostelecky B, Lippincott-Schwartz J. Rambold AS, et al. Autophagy. 2011 Dec;7(12):1568-9. doi: 10.4161/auto.7.12.17992. Autophagy. 2011. PMID: 22024745 Free PMC article. - Autophagy in alcohol-induced liver diseases.
Dolganiuc A, Thomes PG, Ding WX, Lemasters JJ, Donohue TM Jr. Dolganiuc A, et al. Alcohol Clin Exp Res. 2012 Aug;36(8):1301-8. doi: 10.1111/j.1530-0277.2012.01742.x. Epub 2012 May 2. Alcohol Clin Exp Res. 2012. PMID: 22551004 Free PMC article. Review. - Pathophysiological role of autophagy: lesson from autophagy-deficient mouse models.
Ichimura Y, Komatsu M. Ichimura Y, et al. Exp Anim. 2011;60(4):329-45. doi: 10.1538/expanim.60.329. Exp Anim. 2011. PMID: 21791873 Review. - Autophagy, Metabolism, and Cancer.
White E, Mehnert JM, Chan CS. White E, et al. Clin Cancer Res. 2015 Nov 15;21(22):5037-46. doi: 10.1158/1078-0432.CCR-15-0490. Clin Cancer Res. 2015. PMID: 26567363 Free PMC article. Review.
Cited by
- Integration of Metabolomics and Transcriptomics Reveal the Mechanism Underlying Accumulation of Flavonols in Albino Tea Leaves.
Zhang Q, Li C, Jiao Z, Ruan J, Liu MY. Zhang Q, et al. Molecules. 2022 Sep 7;27(18):5792. doi: 10.3390/molecules27185792. Molecules. 2022. PMID: 36144526 Free PMC article. - Combining small-volume metabolomic and transcriptomic approaches for assessing brain chemistry.
Knolhoff AM, Nautiyal KM, Nemes P, Kalachikov S, Morozova I, Silver R, Sweedler JV. Knolhoff AM, et al. Anal Chem. 2013 Mar 19;85(6):3136-43. doi: 10.1021/ac3032959. Epub 2013 Mar 1. Anal Chem. 2013. PMID: 23409944 Free PMC article. - Tumor suppression in mice lacking GABARAP, an Atg8/LC3 family member implicated in autophagy, is associated with alterations in cytokine secretion and cell death.
Salah FS, Ebbinghaus M, Muley VY, Zhou Z, Al-Saadi KR, Pacyna-Gengelbach M, O'Sullivan GA, Betz H, König R, Wang ZQ, Bräuer R, Petersen I. Salah FS, et al. Cell Death Dis. 2016 Apr 28;7(4):e2205. doi: 10.1038/cddis.2016.93. Cell Death Dis. 2016. PMID: 27124579 Free PMC article. - RIPK1 regulates survival of human melanoma cells upon endoplasmic reticulum stress through autophagy.
Luan Q, Jin L, Jiang CC, Tay KH, Lai F, Liu XY, Liu YL, Guo ST, Li CY, Yan XG, Tseng HY, Zhang XD. Luan Q, et al. Autophagy. 2015;11(7):975-94. doi: 10.1080/15548627.2015.1049800. Autophagy. 2015. PMID: 26018731 Free PMC article. - Comprehensive Isotopic Targeted Mass Spectrometry: Reliable Metabolic Flux Analysis with Broad Coverage.
Shi X, Xi B, Jasbi P, Turner C, Jin Y, Gu H. Shi X, et al. Anal Chem. 2020 Sep 1;92(17):11728-11738. doi: 10.1021/acs.analchem.0c01767. Epub 2020 Aug 10. Anal Chem. 2020. PMID: 32697570 Free PMC article.
References
- Kuma A, Mizushima N. Semin Cell Dev Biol. 2010;21:683. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- RC1 CA147961/CA/NCI NIH HHS/United States
- RC1 CA147961-02/CA/NCI NIH HHS/United States
- R37 CA053370-19/CA/NCI NIH HHS/United States
- R01 CA130893/CA/NCI NIH HHS/United States
- R01 CA130893-03/CA/NCI NIH HHS/United States
- R37 CA053370/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources