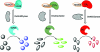Characterization of selective ubiquitin and ubiquitin-like protease inhibitors using a fluorescence-based multiplex assay format - PubMed (original) (raw)
Comparative Study
Characterization of selective ubiquitin and ubiquitin-like protease inhibitors using a fluorescence-based multiplex assay format
Xufan Tian et al. Assay Drug Dev Technol. 2011 Apr.
Abstract
The reversible conjugation of ubiquitin and ubiquitin-like (UbL) proteins to protein substrates plays a critical role in the regulation of many cellular pathways. The removal of ubiquitin from target proteins is performed by ubiquitin proteases also known as deubiquitylases (DUBs). Owing to their substrate specificity and the central role ubiquitylation plays in cell signaling pathways, DUB are attractive targets for therapeutic development. The development of DUB inhibitors requires assays that are amenable to high-throughput screening and provide rapid assessment of inhibitor selectivity. Determination of inhibitor selectivity at an early stage of drug discovery will reduce drug failure in the clinic as well as reduce overall drug development costs. We have developed two novel assays, UbL-Enterokinase light chain and UbL-Granzyme B, for quantifying ubiquitin and UbL protease activity. In our quest to discover and characterize novel chemical entities, we have combined these assays with a previously developed assay in a multiplex format. This multiplex format allows for the detection of three distinct protease activities simultaneously, in a single well. We have demonstrated that the multiplex format is able to distinguish between selective and nonselective protease inhibitors. Specifically, we have used this assay format to characterize P022077, a selective ubiquitin-specific protease 7 inhibitor discovered at Progenra.
Figures
Fig. 1.
Detection of SENP2core protease activity with SUMO3-GZMB and SUMO3-EKL. (A) Increasing concentrations of SENP2core (0 nM ◊, 156 pM ♦, 312 pM □, 625 pM ▪, 1.25 nM ○, 2.5 nM ∙) were incubated with 100 nM SUMO3-EKL, 20 nM of EKL substrate I. The data presented are means ± SEM of triplicate wells. (B) Increasing concentrations of SENP2core (0 nM ◊, 6.25 nM ♦, 12.5 nM □, 25 nM ▪, 50 nM ○, 100 nM ∙) were incubated with 100 nM SUMO3-GZMB and 5 μM IEPD-AMC. The data presented are means ± SEM of triplicate wells. (C) Sub-picomolar concentrations of SENP2core (0 fM ♦, 0.1 fM □, 1 fM ▪, 10 fM ○, 100 fM ∙) were incubated with 500 nM SUMO3-EKL and 200 nM EKL substrate I. Data shown are representative mean data from three independent experiments. AMC, 7-amino-4-methylcoumarin; EKL, enterokinase light chain; GZMB, granzyme B.
Fig. 2.
Schematic representation of multiplexing format. To measure the activity of all three reporters in a single well, three isopeptidases are added to SUMO3-GZMB, ubiquitin-PLA2, and NEDD8-EKL. Each well contains GZMB substrate (ex340/em460), PLA2 substrate (ex460/em538) and EKL substrate II (ex540/em590). PLA2, phospholipase A2. Color images available online at
Fig. 3.
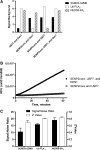
Detection of ubiquitin/ubiquitin-like protease activity in a multiplex assay format. (A) SUMO3-GZMB, ubiquitin-PLA2, and NEDD8-EKL were incubated with either the complete mixture containing SENP2core, USP7, and DEN1, or a mixture lacking one of the three isopeptidases. Fluorescence intensity was measured following a 60-minute incubation and signal:background was calculated. Representative data from one of three independent experiments are shown. Bars represent the signal:background generated by the SUMO3-GZMB reporter (solid bars), ubiquitin-PLA2 reporter (open bars), or the NEDD8-EKL reporter (crosshatched bars) (B) A cocktail of isopeptidases containing USP7 and SENP2core with (closed circles) or without (open circles) DEN1 was added to wells containing SUMO3-GZMB, IEPD-AMC, ubiquitin-PLA2, NBD C6-HPC, NEDD8-EKL, and EKL substrate II. Fluorescence intensity (ex531/em590) was measured as described in the Materials and Methods section. Error bars represent standard error of n = 3 values. (C) Utilizing the data from B, the signal:noise ratio (solid bars) and Z′ ratio (open bars) were calculated as described in Materials and Methods. The data shown are the average of three independent experiments, and the error bars represent the standard error of these three values. NBD C6-HPC, 2-(6-(7-nitrobenz-2-oxa-1,3-diazol-4-yl)amino)hexanoyl-1-hexadecanoyl-_sn_-glycero-3-phosphocholine; USP, ubiquitin-specific protease.
Fig. 4.
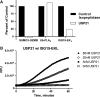
USP21 is a deISGylase as well as a deubiquitylase. (A) 100 nM USP21 was incubated with 100 nM SUMO3-GZMB, 5 μM IEPD-AMC, 100 nM ubiquitin-PLA2, 20 μM NBD C6-HPC, 100 nM SUMO3-EKL, and 1 μM EKL substrate II. Fluorescence intensity was measured after a 60-minute incubation. USP21 deSUMOylase, deubiquitylase, and deISGylase activity were reported relative to control deSUMOylase (20 nM SENP2 core), deubiquitylase (20 nM USP7), and deISGylase (20 nM PLPro) activity, respectively (solid bars). The signal generated by USP21 is represented by open bars. Representative data from three independent experiments is shown. (B) Various concentrations of USP21 (0 nM □, 5 nM ▪, 20 nM ○, 80 nM ∙) were incubated with 50 nM ISG15-EKL and 20 nM EKL substrate I.
Fig. 5.
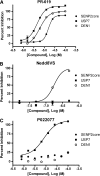
The multiplex format has the utility to identify selective isopeptidase inhibitors. (A) Various concentrations of PR-619 were incubated with 100 nM SENP2core, 20 nM USP7, and 20 nM DEN1. After 30 min, 100 nM SUMO3-GZMB, 5 μM IEPD-AMC, 100 nM ubiquitin-PLA2, 20 μM NBD C6-HPC, 100 nM NEDD8-EKL, and 1 μM EKL Substrate II were added. Fluorescence intensity was measured after 45 min at the appropriate wavelengths and the percent inhibition for each enzyme was calculated. Percent inhibition of each isopeptidase (SENP2core ∙, USP7 ▪, DEN1 ○) is shown plotted versus log of the molar concentration of compound. IC50 values were calculated using GraphPad Prism and are shown where appropriate. (B) A range of concentrations of NEDD8-vinyl sulfone were tested as described above. (C) The selective USP7 inhibitor, P022077, was tested as above.
Similar articles
- Bioluminescence assay platform for selective and sensitive detection of Ub/Ubl proteases.
Orcutt SJ, Wu J, Eddins MJ, Leach CA, Strickler JE. Orcutt SJ, et al. Biochim Biophys Acta. 2012 Nov;1823(11):2079-86. doi: 10.1016/j.bbamcr.2012.06.004. Epub 2012 Jun 15. Biochim Biophys Acta. 2012. PMID: 22705352 Free PMC article. - The Ubiquitin/UBL Drug Target Repertoire.
Osborne HC, Irving E, Schmidt CK. Osborne HC, et al. Trends Mol Med. 2020 Dec;26(12):1133-1134. doi: 10.1016/j.molmed.2020.08.009. Epub 2020 Sep 24. Trends Mol Med. 2020. PMID: 32981839 No abstract available. - Characterization of ubiquitin and ubiquitin-like-protein isopeptidase activities.
Nicholson B, Leach CA, Goldenberg SJ, Francis DM, Kodrasov MP, Tian X, Shanks J, Sterner DE, Bernal A, Mattern MR, Wilkinson KD, Butt TR. Nicholson B, et al. Protein Sci. 2008 Jun;17(6):1035-43. doi: 10.1110/ps.083450408. Epub 2008 Apr 18. Protein Sci. 2008. PMID: 18424514 Free PMC article. - Substrate specificity of the ubiquitin and Ubl proteases.
Ronau JA, Beckmann JF, Hochstrasser M. Ronau JA, et al. Cell Res. 2016 Apr;26(4):441-56. doi: 10.1038/cr.2016.38. Epub 2016 Mar 25. Cell Res. 2016. PMID: 27012468 Free PMC article. Review. - In the moonlight: non-catalytic functions of ubiquitin and ubiquitin-like proteases.
Campos Alonso M, Knobeloch KP. Campos Alonso M, et al. Front Mol Biosci. 2024 Feb 22;11:1349509. doi: 10.3389/fmolb.2024.1349509. eCollection 2024. Front Mol Biosci. 2024. PMID: 38455765 Free PMC article. Review.
Cited by
- Screening of a Focused Ubiquitin-Proteasome Pathway Inhibitor Library Identifies Small Molecules as Novel Modulators of Botulinum Neurotoxin Type A Toxicity.
Sen E, Kota KP, Panchal RG, Bavari S, Kiris E. Sen E, et al. Front Pharmacol. 2021 Sep 27;12:763950. doi: 10.3389/fphar.2021.763950. eCollection 2021. Front Pharmacol. 2021. PMID: 34646144 Free PMC article. - Creating and screening natural product libraries.
Wilson BAP, Thornburg CC, Henrich CJ, Grkovic T, O'Keefe BR. Wilson BAP, et al. Nat Prod Rep. 2020 Jul 1;37(7):893-918. doi: 10.1039/c9np00068b. Epub 2020 Mar 18. Nat Prod Rep. 2020. PMID: 32186299 Free PMC article. Review. - Emerging role of DUBs in tumor metastasis and apoptosis: Therapeutic implication.
He M, Zhou Z, Wu G, Chen Q, Wan Y. He M, et al. Pharmacol Ther. 2017 Sep;177:96-107. doi: 10.1016/j.pharmthera.2017.03.001. Epub 2017 Mar 6. Pharmacol Ther. 2017. PMID: 28279784 Free PMC article. Review. - A small molecule inhibitor of ubiquitin-specific protease-7 induces apoptosis in multiple myeloma cells and overcomes bortezomib resistance.
Chauhan D, Tian Z, Nicholson B, Kumar KG, Zhou B, Carrasco R, McDermott JL, Leach CA, Fulcinniti M, Kodrasov MP, Weinstock J, Kingsbury WD, Hideshima T, Shah PK, Minvielle S, Altun M, Kessler BM, Orlowski R, Richardson P, Munshi N, Anderson KC. Chauhan D, et al. Cancer Cell. 2012 Sep 11;22(3):345-58. doi: 10.1016/j.ccr.2012.08.007. Cancer Cell. 2012. PMID: 22975377 Free PMC article. - Identification of Inhibitors of SARS-CoV-2 3CL-Pro Enzymatic Activity Using a Small Molecule in Vitro Repurposing Screen.
Kuzikov M, Costanzi E, Reinshagen J, Esposito F, Vangeel L, Wolf M, Ellinger B, Claussen C, Geisslinger G, Corona A, Iaconis D, Talarico C, Manelfi C, Cannalire R, Rossetti G, Gossen J, Albani S, Musiani F, Herzog K, Ye Y, Giabbai B, Demitri N, Jochmans D, Jonghe S, Rymenants J, Summa V, Tramontano E, Beccari AR, Leyssen P, Storici P, Neyts J, Gribbon P, Zaliani A. Kuzikov M, et al. ACS Pharmacol Transl Sci. 2021 Mar 11;4(3):1096-1110. doi: 10.1021/acsptsci.0c00216. eCollection 2021 Jun 11. ACS Pharmacol Transl Sci. 2021. PMID: 35287429 Free PMC article.
References
- Schwartz AL. Ciechanover A. Targeting proteins for destruction by the ubiquitin system: implications for human pathobiology. Annu Rev Pharmacol Toxicol. 2009;49:73–96. - PubMed
- Haas AL. Structural insights into early events in the conjugation of ubiquitin and ubiquitin-like proteins. Mol Cell. 2007;27:174–175. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials