Expanding role of ubiquitination in NF-κB signaling - PubMed (original) (raw)
Review
Expanding role of ubiquitination in NF-κB signaling
Siqi Liu et al. Cell Res. 2011 Jan.
Abstract
Best known for its role in targeting protein degradation by the proteasome, ubiquitin modification has also emerged as an important mechanism that regulates cell signaling through proteasome-independent mechanisms. The role of ubiquitin as a versatile signaling tag is characteristically illustrated in the NF-κB pathways, which regulate a variety of physiological and pathological processes in response to diverse stimuli. Here, we review the role of ubiquitination in different steps of the NF-κB signaling cascades, focusing on recent advances in understanding the mechanisms of protein kinase activation by polyubiquitin chains in different pathways that converge on NF-κB.
Figures
Figure 1
Regulation of protein functions by ubiquitination. (A) Schematic representation of the three-step ubiquitination cascade. Monoubiquitination and K63 polyubiquitination generally serve non-proteolytic functions, whereas polyubiquitin chains of other linkages target proteins for degradation by the proteasome. (B) Structure of ubiquitin (PDB code: 1UBQ), highlighting its seven lysine residues.
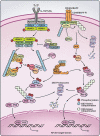
Figure 2
Role of ubiquitination in the canonical and noncanonical NF-κB pathways. In canonical NF-κB activation represented by IL-1R/TLR pathways (left), the receptors recruit the adaptor protein MyD88, the kinases IRAK4 and IRAK1 and the E3 ubiquitin ligase TRAF6. The recruitment causes TRAF6 oligomerization and activates its E3 ligase activity. Together with the E2 enzyme complex Ubc13/Uev1A, TRAF6 synthesizes unanchored K63-polyubiquitin chains that bind to the TAB2 and TAB3 subunits of the TAK1 kinase complex and the NEMO subunit of the IKK complex. This binding brings the kinases into proximity for phosphorylation and activation. IKK subsequently phosphorylates IκBα. Phosphorylated IκBα is recognized by the βTrCP E3 complex and targeted for ubiquitin-mediated proteasomal degradation, releasing the NF-κB dimer p50/p65 into the nucleus to turn on target genes including the IKK inhibitor A20. A20 forms a ubiquitin-editing complex with ITCH, RNF11 and TAX1BP1. One of the functions of this complex is to prevent K63 polyubiquitin chain synthesis by TRAF6 and Ubc13, which leads to IKK inhibition. In the noncanonical pathways (right), stimulation of the BAFF-R and CD40 on the B-cell surface leads to the recruitment of several E3 ligases, including TRAF2, TRAF3 and cIAPs. TRAF2 catalyzes K63 polyubiquitination of cIAPs, which in turn target TRAF3 for degradation by promoting its K48 polyubiquitination. In the absence of TRAF3, NIK is stabilized, leading to the activation of IKKα, which phosphorylates the NF-κB precursor p100. Phosphorylated p100 is also recognized by the βTrCP E3 complex and targeted for ubiquitin-mediated proteasomal processing to form the mature subunit p52. p52 forms a complex with Rel-B, which enters the nucleus to turn on genes that are important for B-cell activation and maturation.
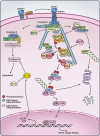
Figure 3
Role of ubiquitination in TNFα-induced NF-κB activation and apoptosis. Binding of TNFα to its receptor (TNFR1) leads to the assembly of a membrane receptor complex (complex I) by recruiting TRADD, TRAF2 or TRAF5, RIP1 and cIAPs. HOIP and HOIL-1 may also help to stabilize complex I by binding to polyubiquitin chains on cIAPs. RIP1 is polyubiquitinated by cIAPs and this modification leads to the recruitment of the TAK1 and IKK complexes. Subsequently, IκBα is phosphorylated and degraded, leading to NF-κB activation. Following complex I formation, TRADD, TRAF2 and RIP1 dissociate from TNF receptor and form a death-inducing complex with FADD and procaspase-8 (complex II) in the cytosol. This allows procaspase-8 to undergo autocatalytic cleavage to generate the mature caspase-8, which initiates apoptosis. However, apoptosis is normally prevented by NF-κB-induced expression of anti-apoptotic proteins such as c-FLIP, which prevents caspase-8 activation. Apoptosis is promoted by ITCH and SMAC, which target c-FLIP and cIAPs, respectively, for proteasomal degradation. NF-κB also induces A20, which forms a ubiquitin-editing complex that targets RIP1 for proteasomal degradation.
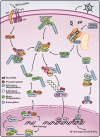
Figure 4
Expanding role of ubiquitination in protein kinase activation in diverse NF-κB pathways. TCR pathway (upper left): Upon TCR stimulation, the serine/threonine kinase PKCθ is activated by a tyrosine kinase cascade. PKCθ promotes the formation of a protein complex composed of CARMA1, BCL10 and MALT1. MALT1 recruits TRAF2 and TRAF6, which catalyze K63 polyubiquitination that leads to the activation of TAK1 and IKK. NLR pathway (upper center): In response to bacterial peptidoglycans, cytosolic receptors NODs recruit TRAF proteins and target RIP2 for K63 ubiquitination, leading to the activation of TAK1 and IKK. RLR pathway (upper right): After viral infection, the viral RNA binds to the cytosolic RNA helicase RIG-I, which then binds to unanchored K63 polyubiquitin chains synthesized by the ubiquitin ligase TRIM25. This binding activates RIG-I, which transduces the signal to MAVS, a mitochondrial membrane protein. MAVS in turn activates IKK and TBK1, leading to the activation of NF-κB and IRF3, respectively. NF-κB and IRF3 enter the nucleus to induce the transcription of type-I interferons (IFN-Is). DNA damage pathway (bottom): Genotoxic stress caused by DNA double-strand breaks triggers the sequential modification of NEMO by SUMO1 and ubiquitin. Monoubiquitinated NEMO exits the nucleus together with ATM and this complex promotes K63 polyubiquitination of ELKS and/or TRAF6, which in turn activates TAK1 and IKK.
Similar articles
- The role of ubiquitin in NF-kappaB regulatory pathways.
Skaug B, Jiang X, Chen ZJ. Skaug B, et al. Annu Rev Biochem. 2009;78:769-96. doi: 10.1146/annurev.biochem.78.070907.102750. Annu Rev Biochem. 2009. PMID: 19489733 Review. - Regulation of NF-κB by ubiquitination.
Chen J, Chen ZJ. Chen J, et al. Curr Opin Immunol. 2013 Feb;25(1):4-12. doi: 10.1016/j.coi.2012.12.005. Epub 2013 Jan 8. Curr Opin Immunol. 2013. PMID: 23312890 Free PMC article. Review. - Ubiquitin signalling in the NF-kappaB pathway.
Chen ZJ. Chen ZJ. Nat Cell Biol. 2005 Aug;7(8):758-65. doi: 10.1038/ncb0805-758. Nat Cell Biol. 2005. PMID: 16056267 Free PMC article. Review. - Diverse roles of the ubiquitin system in NF-κB activation.
Iwai K. Iwai K. Biochim Biophys Acta. 2014 Jan;1843(1):129-36. doi: 10.1016/j.bbamcr.2013.03.011. Epub 2013 Mar 21. Biochim Biophys Acta. 2014. PMID: 23523932 Review. - TRIM13 regulates ubiquitination and turnover of NEMO to suppress TNF induced NF-κB activation.
Tomar D, Singh R. Tomar D, et al. Cell Signal. 2014 Dec;26(12):2606-13. doi: 10.1016/j.cellsig.2014.08.008. Epub 2014 Aug 23. Cell Signal. 2014. PMID: 25152375
Cited by
- A global DNA methylation and gene expression analysis of early human B-cell development reveals a demethylation signature and transcription factor network.
Lee ST, Xiao Y, Muench MO, Xiao J, Fomin ME, Wiencke JK, Zheng S, Dou X, de Smith A, Chokkalingam A, Buffler P, Ma X, Wiemels JL. Lee ST, et al. Nucleic Acids Res. 2012 Dec;40(22):11339-51. doi: 10.1093/nar/gks957. Epub 2012 Oct 16. Nucleic Acids Res. 2012. PMID: 23074194 Free PMC article. - The coactivator role of histone deacetylase 3 in IL-1-signaling involves deacetylation of p65 NF-κB.
Ziesché E, Kettner-Buhrow D, Weber A, Wittwer T, Jurida L, Soelch J, Müller H, Newel D, Kronich P, Schneider H, Dittrich-Breiholz O, Bhaskara S, Hiebert SW, Hottiger MO, Li H, Burstein E, Schmitz ML, Kracht M. Ziesché E, et al. Nucleic Acids Res. 2013 Jan 7;41(1):90-109. doi: 10.1093/nar/gks916. Epub 2012 Oct 19. Nucleic Acids Res. 2013. PMID: 23087373 Free PMC article. - Polyubiquitin-sensor proteins reveal localization and linkage-type dependence of cellular ubiquitin signaling.
Sims JJ, Scavone F, Cooper EM, Kane LA, Youle RJ, Boeke JD, Cohen RE. Sims JJ, et al. Nat Methods. 2012 Feb 5;9(3):303-9. doi: 10.1038/nmeth.1888. Nat Methods. 2012. PMID: 22306808 Free PMC article. - The Role of Nrf2 in Pulmonary Fibrosis: Molecular Mechanisms and Treatment Approaches.
Wang Y, Wei J, Deng H, Zheng L, Yang H, Lv X. Wang Y, et al. Antioxidants (Basel). 2022 Aug 29;11(9):1685. doi: 10.3390/antiox11091685. Antioxidants (Basel). 2022. PMID: 36139759 Free PMC article. Review. - Uev1A promotes breast cancer cell survival and chemoresistance through the AKT-FOXO1-BIM pathway.
Wu Z, Niu T, Xiao W. Wu Z, et al. Cancer Cell Int. 2019 Dec 9;19:331. doi: 10.1186/s12935-019-1050-4. eCollection 2019. Cancer Cell Int. 2019. PMID: 31827405 Free PMC article.
References
- Finley D, Ozkaynak E, Varshavsky A. The yeast polyubiquitin gene is essential for resistance to high temperatures, starvation, and other stresses. Cell. 1987;48:1035–1046. - PubMed
- Pickart CM. Mechanisms underlying ubiquitination. Annu Rev Biochem. 2001;70:503–533. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01-GM63692/GM/NIGMS NIH HHS/United States
- HHMI/Howard Hughes Medical Institute/United States
- R01 GM063692-10/GM/NIGMS NIH HHS/United States
- R01 GM063692-09/GM/NIGMS NIH HHS/United States
- R01 GM063692/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources