Expression of ribosomal protein L22e family members in Drosophila melanogaster: rpL22-like is differentially expressed and alternatively spliced - PubMed (original) (raw)
Expression of ribosomal protein L22e family members in Drosophila melanogaster: rpL22-like is differentially expressed and alternatively spliced
Michael G Kearse et al. Nucleic Acids Res. 2011 Apr.
Abstract
Several ribosomal protein families contain paralogues whose roles may be equivalent or specialized to include extra-ribosomal functions. RpL22e family members rpL22 and rpL22-like are differentially expressed in Drosophila melanogaster: rpL22-like mRNA is gonad specific whereas rpL22 is expressed ubiquitously, suggesting distinctive paralogue functions. To determine if RpL22-like has a divergent role in gonads, rpL22-like expression was analysed by qRT-PCR and western blots, respectively, showing enrichment of rpL22-like mRNA and a 34 kDa (predicted) protein in testis, but not in ovary. Immunohistochemistry of the reproductive tract corroborated testis-specific expression. RpL22-like detection in 80S/polysome fractions from males establishes a role for this tissue-specific paralogue as a ribosomal component. Unpredictably, expression profiles revealed a low abundant, alternative mRNA variant (designated 'rpL22-like short') that would encode a novel protein lacking the C-terminal ribosomal protein signature but retaining part of the N-terminal domain. This variant results from splicing of a retained intron (defined by non-canonical splice sites) within rpL22-like mRNA. Polysome association and detection of a low abundant 13.5 kDa (predicted) protein in testis extracts suggests variant mRNA translation. Collectively, our data show that alternative splicing of rpL22-like generates structurally distinct protein products: ribosomal component RpL22-like and a novel protein with a role distinct from RpL22-like.
Figures
Figure 1.
(A) Clustal W alignment of D. melanogaster RpL22 and RpL22-like sequences. Aligned sequences (RpL22: FBgn0015288; RpL22-like: FBgn0034837) show conservation at the C-terminus (46% sequence identity; 73% sequence similarity), but divergence at the N-terminus (sequence identity 36%; sequence similarity 47%). Overall amino acid identity is 37% (6). Shaded amino acid residues mark conservation within the RpL22e superfamily. Other residues define the N-terminal extension with homology to histone H1 [(7) for RpL22 and RpL23a]. Boxes highlight functional residues involved in nuclear localization (box 1), rRNA binding (boxes 2 and 3) and nucleolar localization (boxes 2 and 3) (20). Underlined residues in the C-termini were used to make polyclonal peptide Abs. (B) Translation of rpL22-like short mRNA (GenBank accession no. HM756190) using Translation Tool (expasy.org) revealed a putative protein of 123 amino acid, consisting primarily of the histone H1-like domain (N-terminus) and not the RpL22-like domain (C-terminus). Underlined residues in the C-termini were used to make polyclonal peptide Abs.
Figure 2.
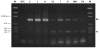
RT-PCR analysis of rpL22-like in different developmental stages, tissues, and in S2 cells. RT-PCR using rpL22-like primers to amplify the coding sequence resulted in the expected 939 bp amplicon (rpL22-like, arrow) in all samples. An additional smaller amplicon (rpL22-like short, arrowhead) of ∼390 bp was present in all samples. Variability in the intensity of rpL22-like short was noted in numerous replicates of the RT-PCR for various samples as noted here for embryonic and adult samples. NTC: no template control; E: embryo; L: larva; A: adult; S2: S2 cells; T: testis; O: ovary; MH: male heads; FH: female heads; M: pGEM marker.
Figure 3.
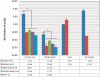
qRT-PCR reveals rpL22-like mRNA enrichment in testis. Using isoform-specific primers, qRT-PCR shows that both isoforms are more highly expressed in testis compared with other tissues. Vasa and β2-tubulin serve as germ cell- and testis-specific controls, respectively. Numbers in table represent _C_T values. *P < 0.01.
Figure 4.
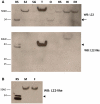
Western blot analysis confirms differential expression of RpL22-like. (A) Compared to the recombinant standard (RS), western analysis for RpL22 shows immunodetection at either the expected MW of 32.9 kDa (arrow) and/or at a higher MW at ∼50 kDa in all tissues. Immunodetection of RpL22-like at the expected MW of 34.3 kDa (arrowhead) is solely visible in testes. Insoluble extracts from male and female heads contain a higher MW species of RpL22-like [whole intact heads of mixed sex with eyes (HI)]. No immunoreactive species is seen in eyeless heads (eyes are dissected out; EH) or in a soluble head extract. The additional lower band in the RS sample is endogenous bacterial protein recognized by mouse antisera (see
Supplementary Figure S2
for additional explanation). (B) The electrophoretic shift in RpL22-like is not sex specific as seen by western analysis of whole head tissue from males (M) and females (F). S2: S2 cells; SG: larval salivary glands; T: testis; O: ovary; HS: soluble head extract.
Figure 5.
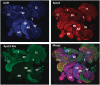
Immunofluorescent staining of RpL22 and RpL22-like in adult male reproductive tract using paralogue-specific Abs. Extruded sperm (ES) were released from the testes during dissection. Spermatogenesis is initiated at the apical tip of the testis (T) and progresses through the testis coils. The most mature sperm are, therefore, located distal to the apical end of the testis. Mature sperm pass from the testis into the seminal vesicle (SV). Seminal fluid is added to sperm from the accessory gland (AG), and sperm are released through the ejaculatory duct (ED). RpL22 (red) is ubiquitously expressed in the reproductive system. RpL22-like (green) is expressed exclusively within the testis and within sperm cells. DNA is visualized by DAPI staining (blue).
Figure 6.
Density gradient ultra-centrifugation showing L22-like association with active translational machinery. Soluble extracts from adult male flies were fractionated on 10–50% sucrose gradients. The top left panel shows the absorbance profile of the sucrose gradient. Extracted RNAs from fractions are shown on the top right. Fractions containing polysomes (poly) and 80S ribosomes were pooled, pelleted separately and subjected to western analysis (bottom panels). Both paralogues were detected at the expected MW in fractions containing 80S ribosomes and polysomes, indicating that both are stable components of translating ribosomes. No sizeable amount of either protein was detected at the top (T) of the gradient. Input: 25 µg and/or 50 µg whole male extract.
Figure 7.
RpL22-like coding region showing novel splice site junctions for rpL22 like-short. Exon sequences (capitalized) and intronic sequences (lowercase) were derived from Flybase (FBgn0034837). The coding region of rpL22-like is shown in blue caps. The previously annotated intron (FlyBase FB2010_06) is shown in green lowercase. The novel intron is represented in bold with non-canonical splice sites underlined (5′SS: CT, 3′SS: CG). The rpL22-like short sequence (GenBank accession no. HM756190) was derived from sequencing multiple (29 total) cloned cDNAs from RT-PCR analyses (Figure 2). Red and purple arrows in the splicing diagram represent primer pairs used in RT-PCR analyses in Figure 8A and
Supplementary Figure S1
.
Figure 8.
(A) RT-PCR analysis of rpL22-like transcripts using primers that bridge exons in rpL22-like and rpL22-like short. RT-PCR products using the exon1/2 bridge primer should hybridize to rpL22-like and rpL22-like short, producing bands of ∼950 bp (arrow) and 392 bp (closed arrowhead), respectively (red primer set in Figure 7). All samples show such products in multiple experiments. Other amplicons (<350 bp) were cloned and sequenced and determined to be the result of non-specific amplification. The novel rpL22-like short exon2/3 bridge primer should specifically hybridize only to rpL22-like short (purple primer set in Figure 7). The expected amplicon of ∼370 bp (open arrowhead) is seen in all samples, confirming the presence of rpL22-like short. E: embryo; L: larval; A: adult; S2: S2 cells; M: pGEM marker. (B) Northern blot analysis of rpL22-like mRNAs from different developmental stages. PolyA+ RNA from embryonic, larval and adult fly stages was probed with 32P-labelled rpL22-like- specific cDNA oligomers. Estimated transcript sizes based on RNA markers are shown in kilobases (kb). rpL22-like mRNA is predicted to be 1.194 kb (Flybase). rpL22-like short mRNA is predicted to be a minimum size of ∼0.625 kb. Arrows highlight prominent transcripts detected in all stages. (C) Embryonic PolyA+RNA was probed with 32P-labelled _rpL22-like-_specific cDNA oligomers [(full specific—lane1) or (flanking—lane 2)]. RNA sizes were determined relative to an RNA marker.
Figure 9.
rpL22-like short mRNA may be translated. (A) Whole male extract was subjected to 10–50% linear sucrose gradient ultra-centrifugation. RT-PCR analysis of gradient fractions representing 80S subunits and polysomes shows association of rpL22-like (arrow) and rpL22-like short (arrowhead) mRNAs with active translating ribosomes, suggesting rpL22-like short is translated. (B) An initial western analysis of 75 µg of testis extract shows immunodetection at the expected MW of ∼34 kDa and lower at ∼25 and ∼13 kDa (faint band; arrow). ‘Short exposure’ was for 3 s and ‘long exposure’ was for 30 min. (C) Increased protein loading to 120 µg of testis extract enhanced immunodetection of the ∼13 kDa band (arrow) with a longer exposure (30 min compared to 4 s for short exposure). Given the abundance of RpL22-like as a ‘sink’ for antiRpL22-like Ab, the membrane was cut to maximize immunodetection of smaller MW proteins (arrowhead). The ∼13 kDa band (arrow) aligns with the recombinant RpL22-like short protein standard. M: pGEM marker; NTC: no template control; E: male extract; B: bottom of gradient; RS: recombinant standard.
Similar articles
- Functional interplay between ribosomal protein paralogues in the eRpL22 family in Drosophila melanogaster.
Mageeney CM, Kearse MG, Gershman BW, Pritchard CE, Colquhoun JM, Ware VC. Mageeney CM, et al. Fly (Austin). 2018;12(3-4):143-163. doi: 10.1080/19336934.2018.1549419. Epub 2018 Nov 29. Fly (Austin). 2018. PMID: 30465696 Free PMC article. - Specialized eRpL22 paralogue-specific ribosomes regulate specific mRNA translation in spermatogenesis in Drosophila melanogaster.
Mageeney CM, Ware VC. Mageeney CM, et al. Mol Biol Cell. 2019 Aug 1;30(17):2240-2253. doi: 10.1091/mbc.E19-02-0086. Epub 2019 Jun 12. Mol Biol Cell. 2019. PMID: 31188709 Free PMC article. - RpL22e, but not RpL22e-like-PA, is SUMOylated and localizes to the nucleoplasm of Drosophila meiotic spermatocytes.
Kearse MG, Ireland JA, Prem SM, Chen AS, Ware VC. Kearse MG, et al. Nucleus. 2013 May-Jun;4(3):241-58. doi: 10.4161/nucl.25261. Epub 2013 Jun 6. Nucleus. 2013. PMID: 23778934 Free PMC article. - Regulatory Roles of Rpl22 in Hematopoiesis: An Old Dog with New Tricks.
Fahl SP, Wang M, Zhang Y, Duc AC, Wiest DL. Fahl SP, et al. Crit Rev Immunol. 2015;35(5):379-400. doi: 10.1615/critrevimmunol.v35.i5.30. Crit Rev Immunol. 2015. PMID: 26853850 Free PMC article. Review. - The other lives of ribosomal proteins.
Bhavsar RB, Makley LN, Tsonis PA. Bhavsar RB, et al. Hum Genomics. 2010 Jun;4(5):327-44. doi: 10.1186/1479-7364-4-5-327. Hum Genomics. 2010. PMID: 20650820 Free PMC article. Review.
Cited by
- Ribosome Structural Changes Dynamically Affect Ribosome Function.
Lindahl L. Lindahl L. Int J Mol Sci. 2024 Oct 17;25(20):11186. doi: 10.3390/ijms252011186. Int J Mol Sci. 2024. PMID: 39456968 Free PMC article. Review. - Diverse somatic Transformer and sex chromosome karyotype pathways regulate gene expression in Drosophila gonad development.
Mahadevaraju S, Pal S, Bhaskar P, McDonald BD, Benner L, Denti L, Cozzi D, Bonizzoni P, Przytycka TM, Oliver B. Mahadevaraju S, et al. bioRxiv [Preprint]. 2024 Aug 12:2024.08.12.607556. doi: 10.1101/2024.08.12.607556. bioRxiv. 2024. PMID: 39372789 Free PMC article. Preprint. - Hydroxylation and translational adaptation to stress: some answers lie beyond the STOP codon.
Katz MJ, Gándara L, De Lella Ezcurra AL, Wappner P. Katz MJ, et al. Cell Mol Life Sci. 2016 May;73(9):1881-93. doi: 10.1007/s00018-016-2160-y. Epub 2016 Feb 13. Cell Mol Life Sci. 2016. PMID: 26874685 Free PMC article. Review. - Functional interplay between ribosomal protein paralogues in the eRpL22 family in Drosophila melanogaster.
Mageeney CM, Kearse MG, Gershman BW, Pritchard CE, Colquhoun JM, Ware VC. Mageeney CM, et al. Fly (Austin). 2018;12(3-4):143-163. doi: 10.1080/19336934.2018.1549419. Epub 2018 Nov 29. Fly (Austin). 2018. PMID: 30465696 Free PMC article. - Specialized eRpL22 paralogue-specific ribosomes regulate specific mRNA translation in spermatogenesis in Drosophila melanogaster.
Mageeney CM, Ware VC. Mageeney CM, et al. Mol Biol Cell. 2019 Aug 1;30(17):2240-2253. doi: 10.1091/mbc.E19-02-0086. Epub 2019 Jun 12. Mol Biol Cell. 2019. PMID: 31188709 Free PMC article.
References
- McIntosh KB, Bonham-Smith PC. Ribosomal protein gene regulation: what about plants? Can. J. Bot. 2006;84:342–362.
- Sugihara Y, Honda H, Iida T, Morinaga T, Hino S, Okajima T, Matsuda T, Nadano D. Proteomic analysis of rodent ribosomes revealed heterogeneity including ribosomal proteins L10-like, L22-like 1, and L39-like. J. Proteome Res. 2010;9:1351–1366. - PubMed
- Kim TY, Ha CW, Huh WK. Differential subcellular localization of ribosomal protein L7 paralogs in Saccharomyces cerevisiae. Mol. Cells. 2009;27:539–546. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous