The Drosophila miR-310 cluster negatively regulates synaptic strength at the neuromuscular junction - PubMed (original) (raw)
Comparative Study
. 2010 Dec 9;68(5):879-93.
doi: 10.1016/j.neuron.2010.11.016.
Karen Tsang, Edward H Liao, Robin Ball, Jay Penney, Jr-Shiuan Yang, Fatima Elazzouzi, Tao He, Athar Chishti, Greg Lnenicka, Eric C Lai, A Pejmun Haghighi
Affiliations
- PMID: 21145002
- PMCID: PMC3034365
- DOI: 10.1016/j.neuron.2010.11.016
Comparative Study
The Drosophila miR-310 cluster negatively regulates synaptic strength at the neuromuscular junction
Kazuya Tsurudome et al. Neuron. 2010.
Abstract
Emerging data implicate microRNAs (miRNAs) in the regulation of synaptic structure and function, but we know little about their role in the regulation of neurotransmission in presynaptic neurons. Here, we demonstrate that the miR-310-313 cluster is required for normal synaptic transmission at the Drosophila larval neuromuscular junction. Loss of miR-310-313 cluster leads to a significant enhancement of neurotransmitter release, which can be rescued with temporally restricted expression of mir-310-313 in larval presynaptic neurons. Kinesin family member, Khc-73 is a functional target for miR-310-313 as its expression is increased in mir-310-313 mutants and reducing it restores normal synaptic function. Cluster mutants show an increase in the active zone protein Bruchpilot accompanied by an increase in electron dense T bars. Finally, we show that repression of Khc-73 by miR-310-313 cluster influences the establishment of normal synaptic homeostasis. Our findings establish a role for miRNAs in the regulation of neurotransmitter release.
Copyright © 2010 Elsevier Inc. All rights reserved.
Figures
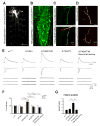
Figure 1. miR-310 cluster is transcribed in larval motor neurons and is a negative regulator of synaptic function
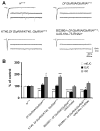
(A–B) Gal4 insertion P{GawB}NP5941 in the miR-310-313 locus drives expression of UAS-mCD8-GFP in (A) CNS, motor axons and peripheral neurons but not in muscles in dissected whole 3rd instar larva (Scale bar is 500 μm) and (B) GFP is detectable in all neurons of the ventral nerve cord at varying degrees of intensity (Scale bar is 50μm) (C) Gal4 insertion P{GawB}NP4255 in the miR-310-313 locus drives expression of UAS-mCD8-GFP in a motor axon innervating muscle 4 in the 3rd abdominal segment (A3) (Scale bar is 10μm). (D) Muscle 4 NMJs of deficiency Df(2R)Exel6070 spanning the miR-310 cluster locus, heterozygote (top) and in trans to KT40 (P{GSV1}GSd033 imprecise excision) (bottom) co-stained with anti-Hrp (green) and anti-Dlg (red) (Scale bar is 10μm). (E) EJPs, EJCs and mEJCs recorded from muscle 6 in the third abdominal segment in wandering third instar larvae in the following genotypes: wild-type (w1118), KT40/+, KT40/KT40, P{GSV1}GSd033 precise excision (KT2/KT2) and BG380/+; KT40/KT40; UAS-miR-310 cluster/+ (KT40 neuronal rescue). For EJPs and EJCs 10 consecutive superimposed traces recorded at 0.5 Hz stimulation rate are shown. For mEJCs sample traces of continuous recording in the absence of stimulation are shown. (F) Quantification for mEJC, EJC and QC normalized to wild-type (w1118) for the indicated genotypes. One-way ANOVA, Games-Howell post hoc test (G) Comparison of QC measurements using Failure analysis at 0.25 mM external [Ca2+]. w1118 (n=6) vs. KT40/KT40 (n=10), P=0.0024. KT2/KT2 (n=7) vs. KT40/KT40 (n=10), P=0.0099. KT40/KT40 vs. KT40/KT40 neuronal (elav-Gal4) rescue (n=8), P=0.0084 One-way ANOVA was used. *P<0.05, **P<0.01 and ***P<0.001. See S7 for details of statistical analysis.
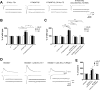
Figure 2. Reduction in Khc-73 leads to suppression of quantal content in KT40 mutants
(A) EJCs and mEJCs for the indicated genotypes. Df(2R)Exel6285 was used as DfKhc-73. UAS-Khc-73-RNAi was driven by the pan-neuronal driver elav-Gal4. (B and C) Quantifications for mEJC, EJC, and QC for the indicated genotypes, n=20, 19, 25, 21 for B and 20, 27, 20, 13 and 20 for C, respectively. One-way ANOVA: *P<0.05, **P<0.01 and ***P<0.001. See S7 for details. (D) EJPs, EJCs and mEJCs for control (BG380-Gal4/+), Khc-73 overexpression (OE) (BG380-Gal4/+;UAS-HA:Khc-73/+) and Khc-73-RNAi overexpression (BG380-Gal4/+;UAS-Khc-73-RNAi/+). (E) Quantification of mEJC, EJC and QC for the genotypes in (D). n=19, 19 and 8, respectively. One-way ANOVA. *P<0.05, **P<0.01 and ***P<0.001. See S7 for details.
Figure 3. Khc-73 is a target of the miR-310 cluster
(A) Schematic of Khc-73 transcript with target sites for the miR-310 cluster indicated. (B) Luciferase activity of vector control, _Khc-73_-3′UTR luciferase reporter (Khc-73 3′UTR), and _Khc-73_-3′UTR with miR-310 cluster sites mutated (Khc-73 3′UTR mutated) in response to the miR-310 cluster (pUAST-miR-310 cluster) and control pUAST-miR-306. ***P=2.02×10−5 Student’s t-test. (C) Luciferase activity of vector control, Khc-73 exon 14 7-mer, Khc-73 exon 14 7-mer mutated and Khc-73 3′UTR in response to miR-310 cluster and miR-306. Khc-73 exon 14 7-mer vs. psiCHECK vector control ***P=2.06×10−5. Khc-73 exon 14 7-mer vs. Khc-73 exon 14 7-mer mutated ***P=0.00125 Student’s t-test. (D) Western blot analysis of Khc-73 protein levels in wild-type (w1118) and KT40 mutants using a Khc-73 specific antibody. Actin is a loading control. (E–L) Motor neuron axon expression of HA:Khc-73 using BG380 Gal4: (E–F) UAS-HA:Khc-73 with Khc-73 3′UTR in (E) Control and (F) KT40/KT40. (G–H) UAS-Khc-73 without Khc-73 3′UTR in (G) Control and (H) KT40/KT40. (I–J) UAS-Khc-73 without Khc-73 3′UTR in (I) Control and (J) miR-310 cluster overexpression. (K–L) UAS-HA:Khc-73 without Khc-73 3′UTR and with exon 14 miR-310 cluster site mutated in (K) Control and (L) miR-310 cluster overexpression. Stained with anti-HA (green) and anti-HRP (red). (Scale is 10μm). See also Figure S3.
Figure 4. miR-310 cluster genetic interactions with Khc-73 and the active zone protein brp
(A) Brp at muscle 4 NMJs in KT40 (middle panel) is increased in intensity compared to wild type control (top panel). Brp staining intensity returns to wild-type in KT40 mutants when Khc-73 is suppressed by RNAi (bottom panel). (B) Quantification of fluorescence intensity of Brp staining normalized to HRP and expressed as a percentage of wild-type. Control (BG380/+) vs. KT40 (BG380/+;KT40/KT40) P=0.0138. KT40 vs. KT40; UAS-Khc-73-RNAi (BG380/+; KT40/KT40; UAS-Khc-73-RNAi/+) P=0.0337. n=12. Student’s t-test. (C) EJCs and mEJCs for the indicated genotypes. Df(2R)Exel6285 was used as DfKhc-73. To induce UAS-Brp expression, first instar larvae were transferred to food containing 50μM RU486. (D) Quantifications for mEJC, EJC, and QC for the indicated genotypes in (C). n=16, 15 and 16, respectively. elav-GS/+ vs. elav-GS/UAS-Brp, P=2.3×10−4 for EJCs and P=7.1×10−5 for QC. elav-GS/UAS-Brp vs. DfKhc-73/+;elav-GS/UAS-Brp, P=9.4×10−4 for EJCs and P=4.8×10−4 for QC. One way ANOVA with Tukey post hoc test was performed for statistical analysis for comparisons (E) Quantifications for mEJC, EJC and QC for the indicated genotypes. Overexpression of UAS-Brp in motor neurons causes a significant increase in QC and the increase in QC in KT40 homozygous larvae was suppressed by removing one copy of Brp. (F) Quantifications for mEJC, EJC and QC for the indicated genotypes. No significant differences found for mEJC, EJC and QC, between larvae overexpressing UAS-Brp in motor neurons (n=15), KT40 homozygotes (n=10) or larvae both homozygote for KT40 and overexpressing UAS-Brp in motor neurons (n=9). One-way ANOVA was performed. See also Figure S4 and S7.
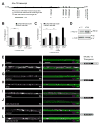
Figure 5. KT40 mutants and Khc-73 overexpression in motor neurons increases the number of T-bars per synaptic density in type Ib boutons
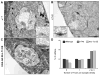
(A) wild-type, (B) KT40/KT40 and (C) Khc-73 overexpression (19 500× magnification, Scale bar 1 μm), Inset images (40 000× magnification, Scale bar is 100nm). (D) KT40 and Khc-73 OE larvae have an increase in the proportion of synaptic densities with two to three T-bars as compared to wild-type. (wild-type n=23, KT40 n=11, and Khc-73 OE n=9 boutons) See also Figure S5.
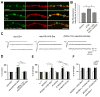
Figure 6. miR-310 cluster shows strong genetic interaction with cacophony calcium channel subunit
(A) EJCs and mEJCs in the indicated genotypes. Removal of one copy of cac (cacHC129) leads to a reduction in the size of EJCs in KT40 mutant larvae. (B) Quantification for mEJC, EJC and QC for the indicated genotypes. * is p<0.05; ** is p<0.01 and ***is p<0.001. One-way ANOVA, Games-Howell (GH) post hoc test. (C) Examples of Ca2+ transients measured in the Ib terminals innervating muscle fiber 6/7 during a single AP in KT40/KT40 and wild-type. The relative increase in [Ca2+]i was determined from the ΔF/F measured at the peak of single APs. (D) Comparison of ΔF/F for KT40 vs control. The relative increase in calcium was measured at the peak of a single AP. (E) Quantification of decay time constants of calcium transients for KT40 and control. There was no significant difference between the mutant and the control. n represents (number of boutons, number of animals). (F) Representative pseudo-color images mapping the ΔF/F during the plateau of a 10 Hz train of APs. Warmer colors represent greater ΔF/F. Calibration bar: 0 to 30 % ΔF/F. Scale is 10 μm. See also Figure S6.
Figure 7. miR-310 cluster and Khc-73 participate in activity dependent synaptic homeostasis
(A) Representative EJCs and mEJCs for the indicated genotypes. DfGluRIIA is Df(2L)clh4. (B) Quantifications for mEJC, EJC, and QC for the indicated genotypes. n=20, 12, 22, 20, 30, 12 and 24, respectively. The increase in QC of GluRIIASP16/DfGluRIIA is suppressed by overexpression of UAS-miR-310 cluster or UAS-Khc-73-RNAi, n=20, 12, 22, 20, 30. One way ANOVA was performed. See S7 for details of statistical analysis. * is p<0.05; ** is p<0.01 and ***is p<0.001.
Similar articles
- The Ly6 neurotoxin-like molecule target of wit regulates spontaneous neurotransmitter release at the developing neuromuscular junction in Drosophila.
Kim NC, Marqués G. Kim NC, et al. Dev Neurobiol. 2012 Dec;72(12):1541-58. doi: 10.1002/dneu.22021. Epub 2012 Jul 27. Dev Neurobiol. 2012. PMID: 22467519 - Mutations in Wnt2 alter presynaptic motor neuron morphology and presynaptic protein localization at the Drosophila neuromuscular junction.
Liebl FL, McKeown C, Yao Y, Hing HK. Liebl FL, et al. PLoS One. 2010 Sep 15;5(9):e12778. doi: 10.1371/journal.pone.0012778. PLoS One. 2010. PMID: 20856675 Free PMC article. - TOR is required for the retrograde regulation of synaptic homeostasis at the Drosophila neuromuscular junction.
Penney J, Tsurudome K, Liao EH, Elazzouzi F, Livingstone M, Gonzalez M, Sonenberg N, Haghighi AP. Penney J, et al. Neuron. 2012 Apr 12;74(1):166-78. doi: 10.1016/j.neuron.2012.01.030. Neuron. 2012. PMID: 22500638 - Transmission, Development, and Plasticity of Synapses.
Harris KP, Littleton JT. Harris KP, et al. Genetics. 2015 Oct;201(2):345-75. doi: 10.1534/genetics.115.176529. Genetics. 2015. PMID: 26447126 Free PMC article. Review. - Synaptic development: insights from Drosophila.
Collins CA, DiAntonio A. Collins CA, et al. Curr Opin Neurobiol. 2007 Feb;17(1):35-42. doi: 10.1016/j.conb.2007.01.001. Epub 2007 Jan 16. Curr Opin Neurobiol. 2007. PMID: 17229568 Review.
Cited by
- Postsynaptic Syntaxin 4 negatively regulates the efficiency of neurotransmitter release.
Harris KP, Littleton JT, Stewart BA. Harris KP, et al. J Neurogenet. 2018 Sep;32(3):221-229. doi: 10.1080/01677063.2018.1501372. Epub 2018 Sep 3. J Neurogenet. 2018. PMID: 30175640 Free PMC article. - A conserved long noncoding RNA affects sleep behavior in Drosophila.
Soshnev AA, Ishimoto H, McAllister BF, Li X, Wehling MD, Kitamoto T, Geyer PK. Soshnev AA, et al. Genetics. 2011 Oct;189(2):455-68. doi: 10.1534/genetics.111.131706. Epub 2011 Jul 20. Genetics. 2011. PMID: 21775470 Free PMC article. - Pervasive Behavioral Effects of MicroRNA Regulation in Drosophila.
Picao-Osorio J, Lago-Baldaia I, Patraquim P, Alonso CR. Picao-Osorio J, et al. Genetics. 2017 Jul;206(3):1535-1548. doi: 10.1534/genetics.116.195776. Epub 2017 May 3. Genetics. 2017. PMID: 28468905 Free PMC article. - miR-153 regulates SNAP-25, synaptic transmission, and neuronal development.
Wei C, Thatcher EJ, Olena AF, Cha DJ, Perdigoto AL, Marshall AF, Carter BD, Broadie K, Patton JG. Wei C, et al. PLoS One. 2013;8(2):e57080. doi: 10.1371/journal.pone.0057080. Epub 2013 Feb 25. PLoS One. 2013. PMID: 23451149 Free PMC article. - A motor neuron protective role of miR-969 mediated by the transcription factor kay.
Zhang X, Lyu J, Jin X, Yang W, Lou Z, Xi Y, Yang X, Ge W. Zhang X, et al. RNA Biol. 2020 Sep;17(9):1277-1283. doi: 10.1080/15476286.2020.1757897. Epub 2020 May 13. RNA Biol. 2020. PMID: 32397794 Free PMC article.
References
- Aravin AA, Lagos-Quintana M, Yalcin A, Zavolan M, Marks D, Snyder B, Gaasterland T, Meyer J, Tuschl T. The Small RNA Profile during Drosophila melanogaster Development. Developmental Cell. 2003;5:337–350. - PubMed
- Ashraf SI, McLoon AL, Sclarsic SM, Kunes S. Synaptic protein synthesis associated with memory is regulated by the RISC pathway in Drosophila. Cell. 2006;124:191–205. - PubMed
- Ball RW, Warren-Paquin M, Tsurudome K, Liao EH, Elazzouzi F, Cavanagh C, An BS, Wang TT, White JH, Haghighi AP. Retrograde BMP signaling controls synaptic growth at the NMJ by regulating trio expression in motor neurons. Neuron. 2010;66:536–549. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous