Epigenetic instability due to defective replication of structured DNA - PubMed (original) (raw)
Epigenetic instability due to defective replication of structured DNA
Peter Sarkies et al. Mol Cell. 2010.
Abstract
The accurate propagation of histone marks during chromosomal replication is proposed to rely on the tight coupling of replication with the recycling of parental histones to the daughter strands. Here, we show in the avian cell line DT40 that REV1, a key regulator of DNA translesion synthesis at the replication fork, is required for the maintenance of repressive chromatin marks and gene silencing in the vicinity of DNA capable of forming G-quadruplex (G4) structures. We demonstrate a previously unappreciated requirement for REV1 in replication of G4 forming sequences and show that transplanting a G4 forming sequence into a silent locus leads to its derepression in REV1-deficient cells. Together, our observations support a model in which failure to maintain processive DNA replication at G4 DNA in REV1-deficient cells leads to uncoupling of DNA synthesis from histone recycling, resulting in localized loss of repressive chromatin through biased incorporation of newly synthesized histones.
Copyright © 2010 Elsevier Inc. All rights reserved.
Figures
Graphical abstract
Figure 1
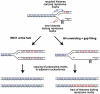
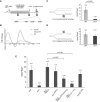
Epigenetic Dysfunction in the β-Globin Locus of rev1 DT40 (A) Map of the region of the chicken β-globin locus studied in this paper (Litt et al., 2001b) between the folate receptor (FR) gene and first of the β-globin genes, ρ. HSA and HS4 are DNase hypersensitive sites that correspond with chromatin domain insulator sequences (Felsenfeld et al., 2004). The distance markers and location of the ChIP primers are indicated below the diagram (see also Table S2). (B) H3K9 dimethylation (H3K9me2) in the WT (solid line) and pcna_K164R (dashed line). In all cases, the specific ChIP signal was normalized to total H3 then to the signal at HS4 (21.37). Error bars represent the standard error of the mean. (C) H3K9 dimethylation (H3K9me2) in rev1 cells derived in our laboratory (solid line, “_rev1 Cambridge”) (Simpson and Sale, 2003) and independently the laboratory of Jean-Marie Buerstedde (dashed line, “rev1 Munich”) (Arakawa et al., 2006). (D) Increased levels of acetylation of the N terminus of H4 in rev1 cells. (E) Acetylation of H3 at K9 and K14 in WT and rev1 cells. (F) Trimethylation of H3 at K4 in WT and rev1 cells. (G) Loss of DNA methylation at the ρ-globin promoter in rev1 cells. Loss of DNA methylation renders the ρ-globin promoter sensitive to restriction by HpaII. Amplification of the ρ-globin promoter by qPCR after HpaII restriction allowed the fraction of DNA remaining uncleaved, and therefore methylated, to be determined. Amplification was normalized to BamHI digested genomic DNA, then further normalized to set the WT level at 1. The amplified region does not contain any BamHI sites. Error bars represent the standard deviation. See also Figure S1.
Figure 2
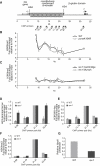
Derepression of ρ-Globin Expression in rev1 Cells (A) Derepression of ρ-globin expression in rev1 cells. Comparison of ρ-globin expression in different DT40 mutants. Expression, monitored by qRT-PCR with primers RhoExpF and R (Table S2), is given as the fold increase over the WT level, which is set at 1. rev1 #162 and #217 are two independent rev1 clones derived in our lab (“rev1 Cambridge”). Error bars show the range. (B) Effect of complementation with human REV1 on ρ-globin derepression. Increase in expression of ρ-globin in rev1 cells, and rev1 cells complemented with hREV1, relative to WT: i, rev1 cells cultured for >3 months; ii, rev1 complemented with hREV1 at 4 weeks, which is as soon as practically possible, and then cultured for >3 months; iii, rev1 cells from i, with established derepression of ρ-globin, complemented with hREV1 and cultured for 3 weeks; iv, as iii, but cultured for 3.5 weeks; and v, as iii, but cultured for 5 weeks. Five weeks in culture corresponds conservatively with 70 cell divisions.
Figure 3
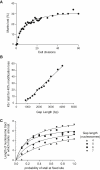
Computational Simulation of Loss of Histone Modifications in Response to Formation of Postreplicative Gaps (A) Percentage of marks lost as a function of time with a fixed stall probability. The graph shows a representative time course for histone modification loss. For this simulation, the postreplicative gap length was set at 1 kb, the probability of two place copy at 0.25 and the probability of fork stalling at 0.025 per nucleosome. This corresponds to one stall every 8 kb. Each data point represents an average of 30 simulations. (B) Postreplicative gap length necessary to produce 40% loss of histone marks in 30 generations. A lower estimate for spontaneous stalling intervals of 60–100kb would clearly place the necessary postreplicative gap length much higher than any current in vivo estimate (see the main text). The x axis scale assumes an internucleosome distance of 200 bp. (C) Length of nucleosome tract in which marks are lost as a function of the probability of stalling per replication cycle at a fixed point. Data is shown for a postreplicative gap length of four to seven nucleosomes, with the variance in length set to 0.
Figure 4
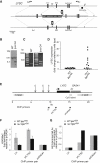
REV1 Is Required for Replication of G Quadruplex-Forming DNA on the Leading-Strand Template (A) Position of G4 DNAs (ovals) in the region of the β-globin locus studied in this work. The sequence of the ρ-globin G4 DNA is shown. (B) Circular dichroism spectroscopy of the ρ-globin G quadruplex forming sequence. Renaturation of the minimal 29 bp G4 oligonucleotide (GQCDG4) in the presence (black line) or absence (light gray line) of K+ ions. Renaturation of a truncated ρ-globin G4 sequence (GQCDG3) in the presence of K+ ions (mid gray line). (C and D)Replication efficiency, shown as the ratio of Ampr to Kanr_E. coli_ colonies, for the ρ-globin G4 DNA on the leading- (C) and lagging- (D) strand template of pQ (Szüts et al., 2008). Error bars represent standard error of the mean. p values were calculated using the unpaired t test (two-tailed). (E) Replication efficiency of the leading-strand template G4 in rev1 mutants. Complementation is with full-length, catalytically inactive (D570AE571A) and C-terminally truncated (1-1137) human REV1. The BRCT mutant is an endogenous deletion of amino acids 69–116 of REV1 (Figure S2). The WT and rev1 data from Figure 4C are shown again for comparison. Error bars represent standard error of the mean. See also Figures S2 and S3.
Figure 5
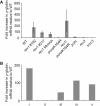
Derepression of Silenced Loci in rev1 Cells Is Associated with G4 DNA (A) Introduction of the ρ-globin G4 DNA into the LYSC locus. The genomic locus was amplified as a SalI (Sa)-ClaI (C) fragment with primers LYSCSalF and LYSCClaR. A linker DNA (G4TplantSph1) containing a BamHI (B) site and the G4 sequence was introduced into the SphI (Sp) site in the first intron of the LYSC gene, so as to be on the feature strand. Correct insertion of the G4 DNA was confirmed by sequencing with primer LYSCG4seq. A bidirectional origin has been demonstrated in the CpG island at the 3′ end of the gene (Phi-van and Strätling, 1999) meaning that the introduced G4 structure will form on the leading-strand template. A puromycin-resistance selection cassette was inserted into the BamHI site. This was then removed by transient expression of Cre recombinase. (B) Southern blot of BamHI-digested DNA showing targeting of one allele of LYSC producing at ∼5 kb band. (C) Confirmation of the presence of the G4 sequence by PCR using primers (LYSCG4F and R) annealing either side of the expected insertion of the G4 DNA (plus the remnants of the loxP recombination sites). (D) qRT-PCR for LYSC expression in clones of WT and rev1 harboring the ρ-globin G4 DNA in the LYSC locus expressed as the fold change relative to unmanipulated WT DT40. (E) Map of the chicken LYSC and GAS41 loci. E, transcription enhancer element; S, transcription suppressor element; MAR, matrix attachment region (adapted from Myers et al., 2003). The positions of the three pairs of ChIP primers are indicated. (F) H3K9 dimethylation (H3K9me2) at the LYSC locus normalized to that at the constitutively active GAS41 promoter in WT and rev1 cells harboring the ρ-globin G4 DNA in one allele of the LYSC locus (lysc+/G4). Error bars represent standard error of the mean. (G) H4 N terminal acetylation at the LYSC locus in WT and rev1 lysc+/G4 cells, normalized to the GAS41 promoter.
Figure 6
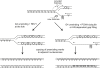
A Model for Loss of Repressive Histone Marks at Sites with G4-Forming Potential in rev1 Cells Replication is depicted arresting at a G4 DNA on the leading-strand template. Parental histones are shown as light-gray circles, with repressive epigenetic marks represented as gray bars. New histones are shown in black. If REV1 is present, the fork can replicate through the G4 DNA, maintaining processive DNA synthesis and histone deposition. It is not clear whether the presence of REV1 prevents the formation of the structure or assists in its unwinding (see also Figure S2). In the absence of REV1 the fork remains arrested at the G4 DNA, resulting in a postreplicative gap. The DNA synthesis associated with the resolution of this gap and of the G4 DNA is accompanied by new histone incorporation resulting in a tract of chromatin lacking the parental epigenetic marks.
Similar articles
- Determinants of G quadruplex-induced epigenetic instability in REV1-deficient cells.
Schiavone D, Guilbaud G, Murat P, Papadopoulou C, Sarkies P, Prioleau MN, Balasubramanian S, Sale JE. Schiavone D, et al. EMBO J. 2014 Nov 3;33(21):2507-20. doi: 10.15252/embj.201488398. Epub 2014 Sep 4. EMBO J. 2014. PMID: 25190518 Free PMC article. - FANCJ coordinates two pathways that maintain epigenetic stability at G-quadruplex DNA.
Sarkies P, Murat P, Phillips LG, Patel KJ, Balasubramanian S, Sale JE. Sarkies P, et al. Nucleic Acids Res. 2012 Feb;40(4):1485-98. doi: 10.1093/nar/gkr868. Epub 2011 Oct 22. Nucleic Acids Res. 2012. PMID: 22021381 Free PMC article. - PrimPol Is Required for Replicative Tolerance of G Quadruplexes in Vertebrate Cells.
Schiavone D, Jozwiakowski SK, Romanello M, Guilbaud G, Guilliam TA, Bailey LJ, Sale JE, Doherty AJ. Schiavone D, et al. Mol Cell. 2016 Jan 7;61(1):161-9. doi: 10.1016/j.molcel.2015.10.038. Epub 2015 Nov 25. Mol Cell. 2016. PMID: 26626482 Free PMC article. - Contributions of the specialised DNA polymerases to replication of structured DNA.
Wickramasinghe CM, Arzouk H, Frey A, Maiter A, Sale JE. Wickramasinghe CM, et al. DNA Repair (Amst). 2015 May;29:83-90. doi: 10.1016/j.dnarep.2015.01.004. Epub 2015 Feb 11. DNA Repair (Amst). 2015. PMID: 25704659 Review. - How is epigenetic information on chromatin inherited after DNA replication?
Nakatani Y, Tagami H, Shestakova E. Nakatani Y, et al. Ernst Schering Res Found Workshop. 2006;(57):89-96. doi: 10.1007/3-540-37633-x_5. Ernst Schering Res Found Workshop. 2006. PMID: 16568950 Review.
Cited by
- The roles of DNA polymerase ζ and the Y family DNA polymerases in promoting or preventing genome instability.
Sharma S, Helchowski CM, Canman CE. Sharma S, et al. Mutat Res. 2013 Mar-Apr;743-744:97-110. doi: 10.1016/j.mrfmmm.2012.11.002. Epub 2012 Nov 26. Mutat Res. 2013. PMID: 23195997 Free PMC article. Review. - Quantitative visualization of DNA G-quadruplex structures in human cells.
Biffi G, Tannahill D, McCafferty J, Balasubramanian S. Biffi G, et al. Nat Chem. 2013 Mar;5(3):182-6. doi: 10.1038/nchem.1548. Epub 2013 Jan 20. Nat Chem. 2013. PMID: 23422559 Free PMC article. - Getting Ready for the Dance: FANCJ Irons Out DNA Wrinkles.
Bharti SK, Awate S, Banerjee T, Brosh RM. Bharti SK, et al. Genes (Basel). 2016 Jul 1;7(7):31. doi: 10.3390/genes7070031. Genes (Basel). 2016. PMID: 27376332 Free PMC article. Review. - Stalled replication forks within heterochromatin require ATRX for protection.
Huh MS, Ivanochko D, Hashem LE, Curtin M, Delorme M, Goodall E, Yan K, Picketts DJ. Huh MS, et al. Cell Death Dis. 2016 May 12;7(5):e2220. doi: 10.1038/cddis.2016.121. Cell Death Dis. 2016. PMID: 27171262 Free PMC article. - The Dynamic Regulation of G-Quadruplex DNA Structures by Cytosine Methylation.
Stevens AJ, de Jong L, Kennedy MA. Stevens AJ, et al. Int J Mol Sci. 2022 Feb 22;23(5):2407. doi: 10.3390/ijms23052407. Int J Mol Sci. 2022. PMID: 35269551 Free PMC article. Review.
References
- Annunziato A.T. Split decision: what happens to nucleosomes during DNA replication? J. Biol. Chem. 2005;280:12065–12068. - PubMed
- Aparicio O., Geisberg J.V., Sekinger E., Yang A., Moqtaderi Z., Struhl K. Chromatin immunoprecipitation for determining the association of proteins with specific genomic sequences in vivo. Curr. Protoc. Mol. Biol. 2005 Chapter 21, Unit 21 23. - PubMed
- Bannister A.J., Zegerman P., Partridge J.F., Miska E.A., Thomas J.O., Allshire R.C., Kouzarides T. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature. 2001;410:120–124. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources