Model of the trimeric fiber and its interactions with the pentameric penton base of human adenovirus by cryo-electron microscopy - PubMed (original) (raw)
Model of the trimeric fiber and its interactions with the pentameric penton base of human adenovirus by cryo-electron microscopy
Hongrong Liu et al. J Mol Biol. 2011.
Abstract
Adenovirus invades host cells by first binding to host receptors through a trimeric fiber, which contains three domains: a receptor-binding knob domain, a long flexible shaft domain, and a penton base-attachment tail domain. Although the structure of the knob domain associated with a portion of the shaft has been solved by X-ray crystallography, the in situ structure of the fiber in the virion is not known; thus, it remains a mystery how the trimeric fiber attaches to its underlying pentameric penton base. By high-resolution cryo-electron microscopy, we have determined the structure of the human adenovirus type 5 (Ad5) to 3.6-Å resolution and have reported the full atomic models for its capsid proteins, but not for the fiber whose density cannot be directly interpreted due to symmetry mismatch with the penton base. Here, we report the determination of the Ad5 fiber structure and its mode of attachment to the pentameric penton base by using an integrative approach of multi-resolution filtering, homology modeling, computational simulation of mismatched symmetries, and fitting of atomic models into cryo-electron microscopy density maps. Our structure reveals that the interactions between the trimeric fiber and the pentameric penton base are mediated by a hydrophobic ring on the top surface of the penton base and three flexible tails inserted into three of the five available grooves formed by neighboring subunits of penton base. These interaction sites provide the molecular basis for the symmetry mismatch and can be targeted for optimizing adenovirus for gene therapy applications.
Copyright © 2010 Elsevier Ltd. All rights reserved.
Figures
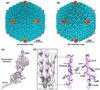
Fig. 1
Overall structure of Ad5 and the penton-base protein. (a–b) Overall structure of Ad5 capsid filtered at 4Å (a) and 10Å (b), respectively, centered on a two-fold axis of icosahedron, showing the fiber (magenta) associating with the penton base (yellow) locate at the vertices of icosahedron. The remaining capsid proteins are colored blue. (c) Superposition of the cryoEM density map (semi-transparent) and the backbone of the atomic model (magenta sticks) of penton-base protein, including the newly resolved ‘N-arm’ region (blue sticks). (d) Representative enlargement of a β sheet [boxed region in (c)] showing separation of strands (left). Stick model of one strand superimposed on its density map (mesh) showing a representative carboxyl (red arrow) density (middle) and is rotated to show side chains (labeled) that are perpendicular to the β sheet (right).
Fig. 2
CryoEM map of the fiber tail and its interaction with the penton base. (a) Top view of the individual penton complex cut from the 4Å density map of Ad5 (Fig. 1a) showing the fiber (magenta) binding to penton base by its tails. The five penton-base monomers are shown in alternating colors cyan, yellow and green. (b) Side view of the fiber density segmented from (a). Five tail densities are visible due to the imposition of five-fold symmetry (left), but two positions (transparent) should be left unoccupied (right). (c) Penton complex showing five fiber-tail densities (magenta) attached to the penton-base pentamer. The fiber shaft density is removed for clarity. Lower inset: Enlargement of the black boxed region showing one fiber tail lying in the groove of two adjacent penton-base monomers. (d) Stereo view of the atomic model (sticks) of an α helix from the top surface of penton-base protein superimposed on its density (mesh). (e) Stereo view of our atomic model (sticks) of the fiber tail (aa7–19) superimposed on its densities (mesh). Three large and distinct side chains (Phe11, Tyr15 and Tyr17) are used as ‘landmarks’ for accurate registration of amino acids. (f) Stereo view of the fiber tail (sticks) inside the groove between two adjacent penton-base monomers (ribbon models, colored yellow and cyan respectively). The interactions between the fiber tail and the penton base include many hydrophobic interactions [side chains Met227, Pro228 and Tyr292 from one penton-base monomer and Leu193, Phe499 and His494 form the other monomer], two probable hydrogen bonds [dashed lines, between Glu203 (penton base) and Asn12 (fiber) and between His495 (penton base) and Tyr15 (fiber)] and one probable salt bridge [dashed lines, between Lys387 (penton base) and Asp18 (fiber)].
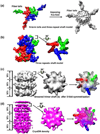
Fig. 3
Fiber model. (a) Ribbon model of the fiber protein (aa319–582) of Ad2 comprises the knob domain and four 15-residue pseudo-repeats in the shaft domain. (b) Sequence alignment and secondary structure prediction for the fiber shaft domain of Ad5. The symbols and colors follow Figure 2 in ref.: the italic types (a–o) indicate the 15-repeat positions; β indicates β-strands; The conserved glycines or prolines in the repeats, residues involved in the hydrophobic and residues possible forming the peripheral hydrophobic patches are filled by purple, yellow and green, respectively. (c) Full space-filling model of Ad5 fiber comprising a global knob, a long shaft and three short N-terminal tails. Three subunits are colored by red, green and blue, respectively.
Fig. 4
Interpreting the fiber density in the cryoEM reconstruction by simulation and fitting. (a) Left: top view of simulated density map of a trimeric fiber. Only three of the 21 repeats of the fiber shaft (same region as the black box in Fig. 3c) are shown. Right: Imposing five-fold symmetry leads to the appearance of five fiber tails and smearing of the fiber shaft. (b) Left: Side view of the first three 15-residue repeats of fiber-shaft model [same as (a), but its tails are removed computationally for clarity]. Right: Surface view of the top slab after being rotated by 90° to show the top view. (c) Same as (b), but after imposing five-fold symmetry. The right panel shows a top slab superimposed by the fiber density before symmetrization (color). (d) CryoEM density map (left) of fiber shaft extracted from the 4Å reconstruction, showing the layered structure (each layer is about 13Å in height, see left panel), and the gear-shaped top slab (middle panel). The right panel shows a superposition of a top slab with the copies of fiber densities before symmetrization (red, green and blue).
Fig. 5
Attachment of the fiber to the penton base revealed at different contour levels. (a–b) Top (a) and side view (b) of the penton complex cut form the whole 10Å map (Fig. 1b), showing the five cable-like densities (‘stay-cables’) connecting the fiber shaft (magenta) to the penton base (yellow). Inset in (b): enlargement of the black box region, showing the fitting of the ribbon models of two penton-base monomers (yellow and red) and a fiber tail (magenta) to the 10Å map (semi-transparent). Also fitting the density map of fiber shaft (magenta) extracted from the 4Å reconstruction. (c) Same as (b), but at different contour level, showing the interactions (black arrows) between the bottom of the fiber shaft and the penton base. (d) Density map of the fiber extracted from (b), showing the ‘stay-cables’ (protrusions) and sites (black arrows) of interaction with the penton base. (e) Ribbon models (same region as the red box in Fig. 2c) showing a hydrophobic ring at the rim of the central pore on the top surface of penton base. Hydrophobic side chains are colored blue. (f) Side view of the penton complex and the enlargement region (inset) showing the fiber tail interacts with the domain of penton base containing an RGD motif.
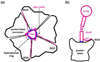
Fig. 6
Schematic illustrations of the attachment of the trimeric fiber (magenta) to the pentameric penton base (black). (a) Top view showing the trimeric fiber attaches to the pentameric penton base at a hydrophobic ring (blue) and three fiber tails (stay-cables). The three sides (a–c) of the magenta triangle denote the three subunits of each fiber. (b) Side view of (a), showing the locations of the hydrophobic interaction (blue), ‘stay-cable’/tail, and the flexible fiber shaft.
Similar articles
- The structure of the human adenovirus 2 penton.
Zubieta C, Schoehn G, Chroboczek J, Cusack S. Zubieta C, et al. Mol Cell. 2005 Jan 7;17(1):121-35. doi: 10.1016/j.molcel.2004.11.041. Mol Cell. 2005. PMID: 15629723 - Atomic structure of human adenovirus by cryo-EM reveals interactions among protein networks.
Liu H, Jin L, Koh SB, Atanasov I, Schein S, Wu L, Zhou ZH. Liu H, et al. Science. 2010 Aug 27;329(5995):1038-43. doi: 10.1126/science.1187433. Science. 2010. PMID: 20798312 Free PMC article. - A quasi-atomic model of human adenovirus type 5 capsid.
Fabry CM, Rosa-Calatrava M, Conway JF, Zubieta C, Cusack S, Ruigrok RW, Schoehn G. Fabry CM, et al. EMBO J. 2005 May 4;24(9):1645-54. doi: 10.1038/sj.emboj.7600653. Epub 2005 Apr 21. EMBO J. 2005. PMID: 15861131 Free PMC article. - Adenovirus structure.
Rux JJ, Burnett RM. Rux JJ, et al. Hum Gene Ther. 2004 Dec;15(12):1167-76. doi: 10.1089/hum.2004.15.1167. Hum Gene Ther. 2004. PMID: 15684694 Review. - Structural Organization and Protein-Protein Interactions in Human Adenovirus Capsid.
Reddy VS, Barry MA. Reddy VS, et al. Subcell Biochem. 2021;96:503-518. doi: 10.1007/978-3-030-58971-4_16. Subcell Biochem. 2021. PMID: 33252742 Review.
Cited by
- Structural Model for Factor X Inhibition of IgM and Complement-Mediated Neutralization of Adenovirus.
Wagner N, Shayakhmetov DM, Stewart PL. Wagner N, et al. Viruses. 2023 Jun 9;15(6):1343. doi: 10.3390/v15061343. Viruses. 2023. PMID: 37376642 Free PMC article. - Latest insights on adenovirus structure and assembly.
San Martín C. San Martín C. Viruses. 2012 May;4(5):847-77. doi: 10.3390/v4050847. Epub 2012 May 21. Viruses. 2012. PMID: 22754652 Free PMC article. Review. - Structures of giant icosahedral eukaryotic dsDNA viruses.
Xiao C, Rossmann MG. Xiao C, et al. Curr Opin Virol. 2011 Aug;1(2):101-9. doi: 10.1016/j.coviro.2011.06.005. Curr Opin Virol. 2011. PMID: 21909343 Free PMC article. Review. - Coxsackievirus and Adenovirus Receptor (CXADR): Recent Findings and Its Role and Regulation in Spermatogenesis.
Zhang Y, Lui WY. Zhang Y, et al. Adv Exp Med Biol. 2021;1288:95-109. doi: 10.1007/978-3-030-77779-1_5. Adv Exp Med Biol. 2021. PMID: 34453733 Review. - Adenovirus Structure: What Is New?
Gallardo J, Pérez-Illana M, Martín-González N, San Martín C. Gallardo J, et al. Int J Mol Sci. 2021 May 15;22(10):5240. doi: 10.3390/ijms22105240. Int J Mol Sci. 2021. PMID: 34063479 Free PMC article. Review.
References
- Roberts DM, Nanda A, Havenga MJE, Abbink P, Lynch DM, Ewald BA, Liu J, Thorner AR, Swanson PE, Gorgone DA, Lifton MA, Lemckert AA, Holterman L, Chen B, Dilraj A, Carville A, Mansfield KG, Goudsmit J, Barouch DH. Hexon-chimaeric adenovirus serotype 5 vectors circumvent pre-existing anti-vector immunity. Nature. 2006;441:239–243. - PubMed
- Everts M, Curiel DT. Transductional targeting of adenoviral cancer gene therapy. Curr. Gene Ther. 2004;4:337–346. - PubMed
- Waddington SN, McVey JH, Bhella D, Parker AL, Barker K, Atoda H, Pink R, Buckley SMK, Greig JA, Denby L, Custers J, Morita T, Francischetti IMB, Monteiro RQ, Barouch DH, Rooijen Nv, Napoli C, Havenga MJE, Nicklin SA, Baker AH. Adenovirus serotype 5 hexon mediates liver gene transfer. Cell. 2008;132:397–409. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- 1S10RR23057/RR/NCRR NIH HHS/United States
- R01 AI069015/AI/NIAID NIH HHS/United States
- GM071940/GM/NIGMS NIH HHS/United States
- CA101904/CA/NCI NIH HHS/United States
- R01 GM071940/GM/NIGMS NIH HHS/United States
- R01 CA101904/CA/NCI NIH HHS/United States
- AI069015/AI/NIAID NIH HHS/United States
- S10 RR023057/RR/NCRR NIH HHS/United States
LinkOut - more resources
Full Text Sources