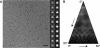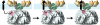Electron cryomicroscopy structure of a membrane-anchored mitochondrial AAA protease - PubMed (original) (raw)
Electron cryomicroscopy structure of a membrane-anchored mitochondrial AAA protease
Sukyeong Lee et al. J Biol Chem. 2011.
Abstract
FtsH-related AAA proteases are conserved membrane-anchored, ATP-dependent molecular machines, which mediate the processing and turnover of soluble and membrane-embedded proteins in eubacteria, mitochondria, and chloroplasts. Homo- and hetero-oligomeric proteolytic complexes exist, which are composed of homologous subunits harboring an ATPase domain of the AAA family and an H41 metallopeptidase domain. Mutations in subunits of mitochondrial m-AAA proteases have been associated with different neurodegenerative disorders in human, raising questions on the functional differences between homo- and hetero-oligomeric AAA proteases. Here, we have analyzed the hetero-oligomeric yeast m-AAA protease composed of homologous Yta10 and Yta12 subunits. We combined genetic and structural approaches to define the molecular determinants for oligomer assembly and to assess functional similarities between Yta10 and Yta12. We demonstrate that replacement of only two amino acid residues within the metallopeptidase domain of Yta12 allows its assembly into homo-oligomeric complexes. To provide a molecular explanation, we determined the 12 Å resolution structure of the intact yeast m-AAA protease with its transmembrane domains by electron cryomicroscopy (cryo-EM) and atomic structure fitting. The full-length m-AAA protease has a bipartite structure and is a hexamer in solution. We found that residues in Yta12, which facilitate homo-oligomerization when mutated, are located at the interface between neighboring protomers in the hexamer ring. Notably, the transmembrane and intermembrane space domains are separated from the main body, creating a passage on the matrix side, which is wide enough to accommodate unfolded but not folded polypeptides. These results suggest a mechanism regarding how proteins are recognized and degraded by m-AAA proteases.
Figures
FIGURE 1.
Protease domains confer hetero-oligomeric assembly of the _m_-AAA protease. A, domain structure of the yeast m_-AAA protease subunits Yta10 (dark gray) and Yta12 (light gray). ND, N-terminal domain; AAA, AAA domain. B, respiratory growth of Δ_yta10_Δ_yta12 cells upon co-expression of Yta10 and chimeras of Yta12. C, respiratory growth of Δ_yta10_Δ_yta12_ cells upon co-expression of Yta12 and chimeras of Yta10. D, forward genetic screen for Yta12 variants allowing respiratory growth in the absence of Yta10. Screening procedures (left panel) and respiratory growth of Δ_yta10_Δ_yta12_ cells upon expression of the identified Yta12 variants (right panel) are shown.
FIGURE 2.
Cryo-EM analysis of an intact yeast _m_-AAA protease. A, representative area of a digital micrograph of the Trap-ATP hexamer embedded in vitreous ice. Selected projection views (right column) and their corresponding class averages (left column) are also shown. Scale bar, 300 Å. B, distribution of particle orientations over an asymmetric unit used in the three-dimensional reconstruction. Brighter dots indicate a larger number of particles.
FIGURE 3.
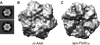
Comparison of the hexameric structures of the _m_-AAA protease and apo-FtsHCyt. A, top-down view of a reference-free two-dimensional class average of the _m_-AAA protease without symmetry (top) and with C6 symmetry applied (bottom). B, 12 Å resolution cryo-EM reconstruction of the _m_-AAA protease in the ATP-bound state depicted as isosurface representation. For clarity, only the matrix domain is shown. C, mass density of the apo-FtsHCyt hexamer calculated at 12 Å resolution from the atomic coordinates of the x-ray structure (PDB code 3KDS (7)).
FIGURE 4.
Cryo-EM structure of an intact yeast _m_-AAA protease. The structure of the Trap-ATP hexamer is shown as isosurface representation with the transmembrane domain colored green, the intermembrane space domain gold, and the matrix domain gray. Because the transmembrane and intermembrane space domains appear to be more flexible than the main body, we split the cryo-EM map into two parts and applied different contour levels. The contour level of the solid isosurface representation was chosen to cover 91 kDa for the transmembrane segments and intermembrane space domains and 436 kDa for the matrix domains. A, top-down view; B, top-down view of the main body only. C, side view; D, vertical central section. The meshed density is contoured at a lower threshold to show the connectivity between the transmembrane segments and the AAA ring.
FIGURE 5.
Atomic structure fit. The cryo-EM reconstruction of the Trap-ATP hexamer is shown as a semitransparent surface with the fitted structure of the hetero-oligomeric _m_-AAA protease docked in. The protease is depicted as ribbon diagram with the transmembrane domain colored green, the AAA red/pink, and the PD blue/sky blue. The dark and light hues represent the Yta10 (red/blue) and Yta12 subunits (pink/sky blue) of the hetero-oligomer. A, top-down views. B, side view (left) and vertical central section (right) of the fitted transmembrane domain, AAA ring, and protease ring. Bound ATP molecules are depicted as CPK models. C, top-down view of the fitted AAA ring. The inset shows an enlarged view of the pore loops and surrounding structural elements. The critical Phe-361 (Yta10) and Phe-421 (Yta12) are depicted as ball-and-stick models. D, model of an Yta12 homo-hexamer. For clarity, three neighboring subunits are colored purple, sky blue, and yellow. Yta12 mutations that allow respiratory growth in the absence of Yta10 are shown as CPK models.
FIGURE 6.
Proposed mechanism of membrane-integral protein dislocation and degradation by the _m_-AAA protease. The _m_-AAA protease hexamer is shown as described in Fig. 4. The mitochondrial inner membrane (blue) and a membrane-integral protein substrate (black) are shown schematically with the initial (star) and secondary contact sites (circle) on the _m_-AAA protease depicted by red symbols.
Similar articles
- Molecular insights into the _m_-AAA protease-mediated dislocation of transmembrane helices in the mitochondrial inner membrane.
Lee S, Lee H, Yoo S, Kim H. Lee S, et al. J Biol Chem. 2017 Dec 8;292(49):20058-20066. doi: 10.1074/jbc.M117.796763. Epub 2017 Oct 13. J Biol Chem. 2017. PMID: 29030426 Free PMC article. - Membrane protein turnover by the m-AAA protease in mitochondria depends on the transmembrane domains of its subunits.
Korbel D, Wurth S, Käser M, Langer T. Korbel D, et al. EMBO Rep. 2004 Jul;5(7):698-703. doi: 10.1038/sj.embor.7400186. Epub 2004 Jun 18. EMBO Rep. 2004. PMID: 15205678 Free PMC article. - Variable and tissue-specific subunit composition of mitochondrial m-AAA protease complexes linked to hereditary spastic paraplegia.
Koppen M, Metodiev MD, Casari G, Rugarli EI, Langer T. Koppen M, et al. Mol Cell Biol. 2007 Jan;27(2):758-67. doi: 10.1128/MCB.01470-06. Epub 2006 Nov 13. Mol Cell Biol. 2007. PMID: 17101804 Free PMC article. - m-AAA proteases, mitochondrial calcium homeostasis and neurodegeneration.
Patron M, Sprenger HG, Langer T. Patron M, et al. Cell Res. 2018 Mar;28(3):296-306. doi: 10.1038/cr.2018.17. Epub 2018 Feb 16. Cell Res. 2018. PMID: 29451229 Free PMC article. Review. - ATP-dependent proteases controlling mitochondrial function in the yeast Saccharomyces cerevisiae.
Van Dyck L, Langer T. Van Dyck L, et al. Cell Mol Life Sci. 1999 Nov 30;56(9-10):825-42. doi: 10.1007/s000180050029. Cell Mol Life Sci. 1999. PMID: 11212342 Free PMC article. Review.
Cited by
- Molecular insights into the _m_-AAA protease-mediated dislocation of transmembrane helices in the mitochondrial inner membrane.
Lee S, Lee H, Yoo S, Kim H. Lee S, et al. J Biol Chem. 2017 Dec 8;292(49):20058-20066. doi: 10.1074/jbc.M117.796763. Epub 2017 Oct 13. J Biol Chem. 2017. PMID: 29030426 Free PMC article. - Unique Structural Features of the Mitochondrial AAA+ Protease AFG3L2 Reveal the Molecular Basis for Activity in Health and Disease.
Puchades C, Ding B, Song A, Wiseman RL, Lander GC, Glynn SE. Puchades C, et al. Mol Cell. 2019 Sep 5;75(5):1073-1085.e6. doi: 10.1016/j.molcel.2019.06.016. Epub 2019 Jul 18. Mol Cell. 2019. PMID: 31327635 Free PMC article. - Folding-Degradation Relationship of a Membrane Protein Mediated by the Universally Conserved ATP-Dependent Protease FtsH.
Yang Y, Guo R, Gaffney K, Kim M, Muhammednazaar S, Tian W, Wang B, Liang J, Hong H. Yang Y, et al. J Am Chem Soc. 2018 Apr 4;140(13):4656-4665. doi: 10.1021/jacs.8b00832. Epub 2018 Mar 21. J Am Chem Soc. 2018. PMID: 29528632 Free PMC article. - Multifunctional Mitochondrial AAA Proteases.
Glynn SE. Glynn SE. Front Mol Biosci. 2017 May 22;4:34. doi: 10.3389/fmolb.2017.00034. eCollection 2017. Front Mol Biosci. 2017. PMID: 28589125 Free PMC article. Review. - Anilinopyrimidine Resistance in Botrytis cinerea Is Linked to Mitochondrial Function.
Mosbach A, Edel D, Farmer AD, Widdison S, Barchietto T, Dietrich RA, Corran A, Scalliet G. Mosbach A, et al. Front Microbiol. 2017 Nov 30;8:2361. doi: 10.3389/fmicb.2017.02361. eCollection 2017. Front Microbiol. 2017. PMID: 29250050 Free PMC article.
References
- Erzberger J. P., Berger J. M. (2006) Annu. Rev. Biophys. Biomol. Struct. 35, 93–114 - PubMed
- Ito K., Akiyama Y. (2005) Annu. Rev. Microbiol. 59, 211–231 - PubMed
- Koppen M., Langer T. (2007) Crit. Rev. Biochem. Mol. Biol. 42, 221–242 - PubMed
- Tatsuta T., Langer T. (2009) Res. Microbiol. 160, 711–717 - PubMed
- Suno R., Niwa H., Tsuchiya D., Zhang X., Yoshida M., Morikawa K. (2006) Mol. Cell 22, 575–585 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases