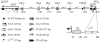Selective interactions of boundaries with upstream region of Abd-B promoter in Drosophila bithorax complex and role of dCTCF in this process - PubMed (original) (raw)
Selective interactions of boundaries with upstream region of Abd-B promoter in Drosophila bithorax complex and role of dCTCF in this process
Olga Kyrchanova et al. Nucleic Acids Res. 2011 Apr.
Abstract
Expression of the genes Ubx, abd-A, and Abd-B of the bithorax complex depends on its cis-regulatory region, which is divided into discrete functional domains (iab). Boundary/insulator elements, named Mcp, Fab-6, Fab-7 and Fab-8 (PTS/F8), have been identified at the borders of the iab domains. Recently, binding sites for a Drosophila homolog of the vertebrate insulator protein CTCF have been identified in Mcp, Fab-6 and Fab-8 and also in several regions that correspond to predicted boundaries, Fab-3 and Fab-4 in particular. Taking into account the inability of the yeast GAL4 activator to stimulate the white promoter when the activator and the promoter are separated by a 5-kb yellow gene, we have tested functional interactions between the boundaries. The results show that all dCTCF-containing boundaries interact with each other. However, inactivation of dCTCF binding sites in Mcp, Fab-6 and PTS/F8 only partially reduces their ability to interact, suggesting the presence of additional protein(s) supporting distant interactions between the boundaries. Interestingly, only Fab-6, Fab-7 (which contains no dCTCF binding sites) and PTS/F8 interact with the upstream region of the Abd-B promoter. Thus, the boundaries might be involved in supporting the specific interactions between iab enhancers and promoters of the bithorax complex.
Figures
Figure 1.
Schemes of the distal part of the bithorax complex including the abd-A and Abd-B loci. The horizontal line represents the bithorax DNA sequence marked off in kilobases according to the coordinates given in (6). The only class A Abd-B transcript that is required for morphogenesis in PS 10 to 13 is drawn above the DNA line. Arrows marked ‘Proximal’ and ‘Distal’ point toward the centromere and the telomere, respectively. Positions of the boundaries and dCTCF-containing regions are indicated by vertical lines. The Abd-B promoter region is shown below the DNA line. Locations of regulatory elements are shown relative to the Abd-B transcription start site (+1). The PTE identified previously (61) is located at –40 relative to the Abd-B transcription start site. The ACTCF region is located at –474 relative to the Abd-B transcription start site. The DNA fragments tested are shown as differently marked boxes. Black circles represent functional binding sites for dCTCF.
Figure 2.
Testing the Fab-3, Fab-4 and Fab-6 elements in the GAL4/white assay. (A) Reductive scheme of transgenic construct used to examine the interaction between regulatory elements at a distance. The yellow and white genes are shown as boxes with arrows indicating the direction of their transcription. Downward arrows indicate target sites for Flp recombinase (frt) or Cre recombinase (lox); the same sites in construct names are denoted by parentheses. GAL4 binding sites (indicated as G4) are at a distance of ∼5 kb from the white promoter. Triangles indicate positions of elements tested for the interaction. (B–D) Experimental evidence that interacting dCTCF-containing regulatory elements facilitate stimulation of white by a distantly located GAL4 activator. Superscript ‘R’ indicates that the corresponding element is inserted in the reverse orientation relative to the white gene in the construct. ‘+ GAL4’ indicates that eye phenotypes in transgenic lines were examined after induction of GAL4 expression. The ‘_white_’ column shows the numbers of transgenic lines with different levels of white pigmentation in the eyes. Wild-type white expression determined the bright red eye color (R); in the absence of white expression, the eyes were white (W). Intermediate levels of pigmentation, with the eye color ranging from pale yellow (pY), through yellow (Y), dark yellow (dY), orange (Or), dark orange (dOr) and brown (Br) to brownish red (BrR), reflect the increasing levels of white expression. N is the number of lines in which flies acquired a new y phenotype upon induction of GAL4 or deletion (Δ) of the specified DNA fragment; T is the total number of lines examined for each particular construct. Other designations are as in Figure 1.
Figure 3.
Testing the functional interaction between regulatory elements containing dCTCF binding sites. For designations, see Figures 1 and 2.
Figure 4.
Role of dCTCF in self-pairing of (A, B) Mcp, (C) Fab-6 and (D–F) PTS/F8 boundaries. The results presented in (A) are from ref. 40. For designations, see Figures 1 and 2.
Figure 5.
Testing the interaction of the Fab-7 boundary with (A) PTS/F8m, (B and C) Fab-3, (D) Fab-4, (E and F) Mcp and (G) Fab-6. For designations, see Figures 1 and 2.
Figure 6.
Analysis of interactions between the ACTCF region of the Abd-B promoter and the Fab-7 and PTS/F8 boundaries. Experiments were performed to compare the interactions of (A) normal ACTCF and (B) mutant ACTCFm with Fab-7. Similar experiments were performed to study the interactions of PTS/F8 with (C) ACTCF and (D) ACTCFm, as well as (E) of PTS/F8m with ACTCF. In addition, ACTCF was tested in combinations with (F and G) the Fab-8 insulator and (H) PTS. The results presented in (C) are from ref. (46). For designations, see Figures 1 and 2.
Figure 7.
Analysis of interaction between the ACTCF region of the Abd-B promoter and (A) Fab-6, (B) mutant Fab-6m, (C and D) Fab-3, (E) Fab-4 and (F–H) Mcp elements. For designations, see Figures 1 and 2.
Similar articles
- Architectural protein Pita cooperates with dCTCF in organization of functional boundaries in Bithorax complex.
Kyrchanova O, Zolotarev N, Mogila V, Maksimenko O, Schedl P, Georgiev P. Kyrchanova O, et al. Development. 2017 Jul 15;144(14):2663-2672. doi: 10.1242/dev.149815. Epub 2017 Jun 15. Development. 2017. PMID: 28619827 Free PMC article. - Fragments of the Fab-3 and Fab-4 Boundaries of the Drosophila melanogaster Bithorax Complex That Include CTCF Sites Are not Effective Insulators.
Kyrchanova OV, Postika NY, Sokolov VV, Georgiev PG. Kyrchanova OV, et al. Dokl Biochem Biophys. 2022 Feb;502(1):21-24. doi: 10.1134/S1607672922010069. Epub 2022 Mar 11. Dokl Biochem Biophys. 2022. PMID: 35275301 Free PMC article. - Functional Dissection of the Blocking and Bypass Activities of the Fab-8 Boundary in the Drosophila Bithorax Complex.
Kyrchanova O, Mogila V, Wolle D, Deshpande G, Parshikov A, Cléard F, Karch F, Schedl P, Georgiev P. Kyrchanova O, et al. PLoS Genet. 2016 Jul 18;12(7):e1006188. doi: 10.1371/journal.pgen.1006188. eCollection 2016 Jul. PLoS Genet. 2016. PMID: 27428541 Free PMC article. - Chromatin domain boundaries in the Bithorax complex.
Mihaly J, Hogga I, Barges S, Galloni M, Mishra RK, Hagstrom K, Müller M, Schedl P, Sipos L, Gausz J, Gyurkovics H, Karch F. Mihaly J, et al. Cell Mol Life Sci. 1998 Jan;54(1):60-70. doi: 10.1007/s000180050125. Cell Mol Life Sci. 1998. PMID: 9487387 Free PMC article. Review. - Unraveling cis-regulatory mechanisms at the abdominal-A and Abdominal-B genes in the Drosophila bithorax complex.
Akbari OS, Bousum A, Bae E, Drewell RA. Akbari OS, et al. Dev Biol. 2006 May 15;293(2):294-304. doi: 10.1016/j.ydbio.2006.02.015. Epub 2006 Mar 20. Dev Biol. 2006. PMID: 16545794 Review.
Cited by
- Stem-loop and circle-loop TADs generated by directional pairing of boundary elements have distinct physical and regulatory properties.
Ke W, Fujioka M, Schedl P, Jaynes JB. Ke W, et al. Elife. 2024 Aug 7;13:RP94114. doi: 10.7554/eLife.94114. Elife. 2024. PMID: 39110491 Free PMC article. - Architectural protein Pita cooperates with dCTCF in organization of functional boundaries in Bithorax complex.
Kyrchanova O, Zolotarev N, Mogila V, Maksimenko O, Schedl P, Georgiev P. Kyrchanova O, et al. Development. 2017 Jul 15;144(14):2663-2672. doi: 10.1242/dev.149815. Epub 2017 Jun 15. Development. 2017. PMID: 28619827 Free PMC article. - Chromosome structure in Drosophila is determined by boundary pairing not loop extrusion.
Bing X, Ke W, Fujioka M, Kurbidaeva A, Levitt S, Levine M, Schedl P, Jaynes JB. Bing X, et al. Elife. 2024 Aug 7;13:RP94070. doi: 10.7554/eLife.94070. Elife. 2024. PMID: 39110499 Free PMC article. - Surviving an identity crisis: a revised view of chromatin insulators in the genomics era.
Matzat LH, Lei EP. Matzat LH, et al. Biochim Biophys Acta. 2014 Mar;1839(3):203-14. doi: 10.1016/j.bbagrm.2013.10.007. Epub 2013 Nov 1. Biochim Biophys Acta. 2014. PMID: 24189492 Free PMC article. Review. - The Functions and Mechanisms of Action of Insulators in the Genomes of Higher Eukaryotes.
Melnikova LS, Georgiev PG, Golovnin AK. Melnikova LS, et al. Acta Naturae. 2020 Oct-Dec;12(4):15-33. doi: 10.32607/actanaturae.11144. Acta Naturae. 2020. PMID: 33456975 Free PMC article.
References
- Maeda RK, Karch F. The bithorax complex of Drosophila: an exceptional Hox cluster. Curr. Top. Dev. Biol. 2009;88:1–33. - PubMed
- Lewis EB. A gene complex controlling segmentation in Drosophila. Nature. 1978;276:565–570. - PubMed
- Sanchez-Herrero E, Vernos I, Marco R, Morato G. Genetic organization of Drosophila bithorax complex. Nature. 1985;313:108–113. - PubMed
- Maeda RK, Karch F. The ABC of the BX-C: the bithorax complex explained. Development. 2006;133:1413–1422. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous