Analysis of streptococcal CRISPRs from human saliva reveals substantial sequence diversity within and between subjects over time - PubMed (original) (raw)
Analysis of streptococcal CRISPRs from human saliva reveals substantial sequence diversity within and between subjects over time
David T Pride et al. Genome Res. 2011 Jan.
Abstract
Viruses may play an important role in the evolution of human microbial communities. Clustered regularly interspaced short palindromic repeats (CRISPRs) provide bacteria and archaea with adaptive immunity to previously encountered viruses. Little is known about CRISPR composition in members of human microbial communities, the relative rate of CRISPR locus change, or how CRISPR loci differ between the microbiota of different individuals. We collected saliva from four periodontally healthy human subjects over an 11- to 17-mo time period and analyzed CRISPR sequences with corresponding streptococcal repeats in order to improve our understanding of the predominant features of oral streptococcal adaptive immune repertoires. We analyzed a total of 6859 CRISPR bearing reads and 427,917 bacterial 16S rRNA gene sequences. We found a core (ranging from 7% to 22%) of shared CRISPR spacers that remained stable over time within each subject, but nearly a third of CRISPR spacers varied between time points. We document high spacer diversity within each subject, suggesting constant addition of new CRISPR spacers. No greater than 2% of CRISPR spacers were shared between subjects, suggesting that each individual was exposed to different virus populations. We detect changes in CRISPR spacer sequence diversity over time that may be attributable to locus diversification or to changes in streptococcal population structure, yet the composition of the populations within subjects remained relatively stable. The individual-specific and traceable character of CRISPR spacer complements could potentially open the way for expansion of the domain of personalized medicine to the oral microbiome, where lineages may be tracked as a function of health and other factors.
Figures
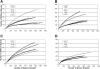
Figure 1.
Rarefaction analysis of CRISPR spacers in the saliva of human subjects at each sampled time point. Rarefaction curves were created using 10,000 random iterations based on spacer richness. (A) Subject #1; (B) subject #2; (C) subject #3; (D) subject #4. (Open circle) Month −6; (open square) Month −3; (closed triangle) Day 1; (closed square) Day 30; (closed circle) Day 60; (open triangle) Month 11.
Figure 2.
Heatmap of unique spacers present in each subject at all time points. Each row represents a unique spacer sequence. The intensity scale bar is located to the right.
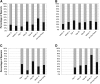
Figure 3.
Shared CRISPR spacers in the saliva of individual human subjects at each time point. (A) Subject #1; (B) subject #2; (C) subject #3; (D) subject #4. (Gray) The proportion of spacers shared with other time points within each subject; (black) spacers that are unique to each time point within each subject.
Figure 4.
Principal coordinates analysis of CRISPR spacer composition from human saliva based on beta diversity. (Gray triangles) Subject #1; (gray squares) subject #2; (black circles) subject #3; (black diamonds) subject #4.
Figure 5.
Heatmap of bacterial OTU abundance based on analysis of 16S rRNA gene sequences from each of the samples and subjects. OTUs were determined by phylogenetic analysis of 16S rRNA sequence alignments, using a 97% cutoff value. Each row represents a unique OTU sequence based on the cutoff criterion. The intensity scale bar is located to the right. Taxonomic labels are shown along the _y_-axis, with OTUs from the genus Streptococcus indicated with a blue brace.
Figure 6.
Principal coordinates analysis of OTU composition based on 16S rRNA gene sequence data from the saliva of each subject. Input to the analysis was beta unweighted unifrac distances. (Gray triangles) Subject #1; (gray squares) subject #2; (black circles) subject #3; (black diamonds) subject #4.
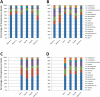
Figure 7.
Streptococcus species in human subject saliva at each time point. Each species is displayed as a percentage of the total number of OTUs identified taxonomically to the Streptococcus genus. (A) Subject #1; (B) subject #2; (C) subject #3; (D) subject #4.
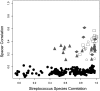
Figure 8.
Pearson correlation scores for comparisons between CRISPR spacer content and streptococcal species composition. Intrasubject comparisons: (open squares) subject #1, (gray triangles) subject #2, (open circles) subject #3, (gray diamonds) subject #4; (black circles) intersubject comparisons.
Figure 9.
Structure of a CRISPR locus from subject #2 at different time points. 2Mut38 represents an isolate of S. sanguinis from subject #2 recovered at Month 11.
Similar articles
- Comparisons of clustered regularly interspaced short palindromic repeats and viromes in human saliva reveal bacterial adaptations to salivary viruses.
Pride DT, Salzman J, Relman DA. Pride DT, et al. Environ Microbiol. 2012 Sep;14(9):2564-76. doi: 10.1111/j.1462-2920.2012.02775.x. Epub 2012 May 15. Environ Microbiol. 2012. PMID: 22583485 Free PMC article. - Diverse CRISPRs evolving in human microbiomes.
Rho M, Wu YW, Tang H, Doak TG, Ye Y. Rho M, et al. PLoS Genet. 2012;8(6):e1002441. doi: 10.1371/journal.pgen.1002441. Epub 2012 Jun 13. PLoS Genet. 2012. PMID: 22719260 Free PMC article. - Conservation of streptococcal CRISPRs on human skin and saliva.
Robles-Sikisaka R, Naidu M, Ly M, Salzman J, Abeles SR, Boehm TK, Pride DT. Robles-Sikisaka R, et al. BMC Microbiol. 2014 Jun 6;14:146. doi: 10.1186/1471-2180-14-146. BMC Microbiol. 2014. PMID: 24903519 Free PMC article. - Clustered regularly interspaced short palindromic repeats (CRISPRs): the hallmark of an ingenious antiviral defense mechanism in prokaryotes.
Al-Attar S, Westra ER, van der Oost J, Brouns SJ. Al-Attar S, et al. Biol Chem. 2011 Apr;392(4):277-89. doi: 10.1515/BC.2011.042. Epub 2011 Feb 7. Biol Chem. 2011. PMID: 21294681 Review. - CRISPR-based adaptive and heritable immunity in prokaryotes.
van der Oost J, Jore MM, Westra ER, Lundgren M, Brouns SJ. van der Oost J, et al. Trends Biochem Sci. 2009 Aug;34(8):401-7. doi: 10.1016/j.tibs.2009.05.002. Epub 2009 Jul 29. Trends Biochem Sci. 2009. PMID: 19646880 Review.
Cited by
- CRISPR-spacer integration reporter plasmids reveal distinct genuine acquisition specificities among CRISPR-Cas I-E variants of Escherichia coli.
Díez-Villaseñor C, Guzmán NM, Almendros C, García-Martínez J, Mojica FJ. Díez-Villaseñor C, et al. RNA Biol. 2013 May;10(5):792-802. doi: 10.4161/rna.24023. Epub 2013 Feb 27. RNA Biol. 2013. PMID: 23445770 Free PMC article. - Biology and genome sequence of Streptococcus mutans phage M102AD.
Delisle AL, Guo M, Chalmers NI, Barcak GJ, Rousseau GM, Moineau S. Delisle AL, et al. Appl Environ Microbiol. 2012 Apr;78(7):2264-71. doi: 10.1128/AEM.07726-11. Epub 2012 Jan 27. Appl Environ Microbiol. 2012. PMID: 22287009 Free PMC article. - Spatial structure and Lamarckian adaptation explain extreme genetic diversity at CRISPR locus.
Haerter JO, Sneppen K. Haerter JO, et al. mBio. 2012 Jul 17;3(4):e00126-12. doi: 10.1128/mBio.00126-12. Print 2012. mBio. 2012. PMID: 22807565 Free PMC article. - CRISPR-based adaptive immune systems.
Terns MP, Terns RM. Terns MP, et al. Curr Opin Microbiol. 2011 Jun;14(3):321-7. doi: 10.1016/j.mib.2011.03.005. Epub 2011 Apr 29. Curr Opin Microbiol. 2011. PMID: 21531607 Free PMC article. Review. - CRISPR dynamics during the interaction between bacteria and phage in the first year of life.
Wu J, Zhang H, Gan R, Xia Y, Zhang F, Wang D, Fu J, Barraclough TG. Wu J, et al. Microb Genom. 2023 Jul;9(7):mgen001053. doi: 10.1099/mgen.0.001053. Microb Genom. 2023. PMID: 37402176 Free PMC article.
References
- Andersson AF, Banfield JF 2008. Virus population dynamics and acquired virus resistance in natural microbial communities. Science 320: 1047–1050 - PubMed
- Barrangou R, Fremaux C, Deveau H, Richards M, Boyaval P, Moineau S, Romero DA, Horvath P 2007. CRISPR provides acquired resistance against viruses in prokaryotes. Science 315: 1709–1712 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources