Nucleosome-mediated cooperativity between transcription factors - PubMed (original) (raw)
Nucleosome-mediated cooperativity between transcription factors
Leonid A Mirny. Proc Natl Acad Sci U S A. 2010.
Abstract
Cooperative binding of transcription factors (TFs) to promoters and other regulatory regions is essential for precise gene expression. The classical model of cooperativity requires direct interactions between TFs, thus constraining the arrangement of TF sites in regulatory regions. Recent genomic and functional studies, however, demonstrate a great deal of flexibility in such arrangements with variable distances, numbers of sites, and identities of TF sites located in cis-regulatory regions. Such flexibility is inconsistent with cooperativity by direct interactions between TFs. Here, we demonstrate that strong cooperativity among noninteracting TFs can be achieved by their competition with nucleosomes. We find that the mechanism of nucleosome-mediated cooperativity is analogous to cooperativity in another multimolecular complex: hemoglobin. This surprising analogy provides deep insights, with parallels between the heterotropic regulation of hemoglobin (e.g., the Bohr effect) and the roles of nucleosome-positioning sequences and chromatin modifications in gene expression. Nucleosome-mediated cooperativity is consistent with several experimental studies, is equally applicable to repressors and activators, allows substantial flexibility in and modularity of regulatory regions, and provides a rationale for a broad range of genomic and evolutionary observations. Striking parallels between cooperativity in hemoglobin and in transcriptional regulation point to a general mechanism that can be used in various biological systems.
Conflict of interest statement
The author declares no conflict of interest.
Figures
Fig. 1.
The model of nucleosome-mediated cooperativity. (A) DNA region containing an array of n sites (green boxes) that can be bound by a histone core (red oval), thus becoming nucleosomal DNA, or remain naked. In either the nucleosomal (N) or the open (O) state, the DNA can be bound by transcription factors (TFs, green ovals). Binding of TFs to the nucleosomal DNA is diminished as compared to naked DNA but is possible due to transient, partial unwinding of the DNA (shown in B) (36). The equilibrium of the system is fully characterized by the scheme in C. The states of the system: nucleosomal (N i) and open (O i), with i being the number of TFs bound. In this form, the nucleosome-TF system is identical to the Monod–Wyman–Changeux (MWC) model of cooperativity in hemoglobin (D). The N and O forms of the DNA correspond to the T and R states of hemoglobin; TF binding is equivalent to O2 binding (see
Table S1
). Like the MWC model, the nucleosome-TF system is determined by three dimensionless parameters: L, c and α (see text). (B) The model of Polach and Widom (19): synergistic binding by two TFs to nearby sites through partial unwinding of nucleosomal DNA. The mechanism requires the constant presence of a nucleosome and does not lead to nucleosome eviction. In our model, binding of multiple TFs can evict a nucleosome completely thus allowing more distant sites to interact, more sites to be involved and hence a higher Hill coefficient.
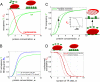
Fig. 2.
Nucleosome-mediated cooperativity and its implications. (A) Cooperative transition in the equilibrium TFBS occupancy, Y (solid green line), and nucleosome occupancy, Y N (red line), as a function of TF concentration α (Eqs. 1 and 2, n = 6, L = 103, c = 10-3). Notice that nucleosome-mediated cooperativity leads to suppression of TF binding at a low concentration, as compared to noncooperative binding (dashed green line). (B) Probability of having at least 3 TFs bound P_3 as a function of TF concentration (blue). Mean occupancy per site, Y, is shown for comparison (green). (C) The Bohr effect: attenuation of histone affinity for DNA, due to modifications or as a function of DNA sequence (modified—dashed line, unmodified—solid line), leads to a shift in TF-nucleosome competition and displacement of the nucleosome by TFs (arrow). This competition renders nucleosomal occupancy, Y N, considerably responsive to small changes in nucleosome affinity (Inset), as demonstrated by the dependence of Y N on -Δ_G = k_B_T log(L/_L_′) (kcal/mol). (D) The effect of number of TF sites, n, on nucleosome stability, obtained for three concentrations of TF: α = 3, 5, 8 (lines from top to bottom). There is a critical number of sites (∼4–5) below which TFs are unable to displace a nucleosome and above which the nucleosome is unstable even at a lower concentration of TFs.
Fig. 3.
Cooperative binding to high- and low-affinity sites. The nucleosomal (red) and TF (green) occupancy profiles for a regulatory region that contains clusters of high- and low-affinity sites. (A) The binding energy profile: a cluster of 8 low-affinity sites and a cluster of 5 high-affinity sites located over the background of spurious low-affinity sites (27). The region contains seven stable, equally spaced nucleosomes with a liner of 50 bp. (B_–_D) Diagrams show nucleosomal occupancy Y N (red), and TF cluster occupancy _P_3 for three values of TF concentration. While an intermediate TF concentration is sufficient to get high-affinity clusters nucleosome-free and TF-bound, a higher concentration is needed for low-affinity clusters. A combination of low- and high-affinity sites in a regulatory region can result in different responses to various TF concentrations. Notice that isolated low-affinity sites are unable to displace nucleosomes. Nucleosomes were assumed to be well-positioned by DNA sequence, and sliding was disregarded. The following parameters were used: c = 0.01; L = 1,000, _α_non-site = 0.001; _α_high-affinity = 20; _α_low-affinity = 1.
Similar articles
- Identifying cooperative transcription factors in yeast using multiple data sources.
Lai FJ, Jhu MH, Chiu CC, Huang YM, Wu WS. Lai FJ, et al. BMC Syst Biol. 2014;8 Suppl 5(Suppl 5):S2. doi: 10.1186/1752-0509-8-S5-S2. Epub 2014 Dec 12. BMC Syst Biol. 2014. PMID: 25559499 Free PMC article. - DNA sequence preferences of transcriptional activators correlate more strongly than repressors with nucleosomes.
Charoensawan V, Janga SC, Bulyk ML, Babu MM, Teichmann SA. Charoensawan V, et al. Mol Cell. 2012 Jul 27;47(2):183-92. doi: 10.1016/j.molcel.2012.06.028. Mol Cell. 2012. PMID: 22841002 Free PMC article. - Cooperative binding between distant transcription factors is a hallmark of active enhancers.
Rao S, Ahmad K, Ramachandran S. Rao S, et al. Mol Cell. 2021 Apr 15;81(8):1651-1665.e4. doi: 10.1016/j.molcel.2021.02.014. Epub 2021 Mar 10. Mol Cell. 2021. PMID: 33705711 Free PMC article. - Primary Role of the Nucleosome.
Kornberg RD, Lorch Y. Kornberg RD, et al. Mol Cell. 2020 Aug 6;79(3):371-375. doi: 10.1016/j.molcel.2020.07.020. Mol Cell. 2020. PMID: 32763226 Review. - Transcription factor access to promoter elements.
Morse RH. Morse RH. J Cell Biochem. 2007 Oct 15;102(3):560-70. doi: 10.1002/jcb.21493. J Cell Biochem. 2007. PMID: 17668451 Review.
Cited by
- Eukaryotic gene regulation at equilibrium, or non?
Zoller B, Gregor T, Tkačik G. Zoller B, et al. Curr Opin Syst Biol. 2022 Sep;31:100435. doi: 10.1016/j.coisb.2022.100435. Epub 2022 Oct 20. Curr Opin Syst Biol. 2022. PMID: 36590072 Free PMC article. - Pluripotency factors determine gene expression repertoire at zygotic genome activation.
Gao M, Veil M, Rosenblatt M, Riesle AJ, Gebhard A, Hass H, Buryanova L, Yampolsky LY, Grüning B, Ulianov SV, Timmer J, Onichtchouk D. Gao M, et al. Nat Commun. 2022 Feb 10;13(1):788. doi: 10.1038/s41467-022-28434-1. Nat Commun. 2022. PMID: 35145080 Free PMC article. - Pou5f3, SoxB1, and Nanog remodel chromatin on high nucleosome affinity regions at zygotic genome activation.
Veil M, Yampolsky LY, Grüning B, Onichtchouk D. Veil M, et al. Genome Res. 2019 Mar;29(3):383-395. doi: 10.1101/gr.240572.118. Epub 2019 Jan 23. Genome Res. 2019. PMID: 30674556 Free PMC article. - Evolution of transcription factor binding through sequence variations and turnover of binding sites.
Krieger G, Lupo O, Wittkopp P, Barkai N. Krieger G, et al. Genome Res. 2022 Jun;32(6):1099-1111. doi: 10.1101/gr.276715.122. Epub 2022 May 26. Genome Res. 2022. PMID: 35618416 Free PMC article. - Complex signal processing in synthetic gene circuits using cooperative regulatory assemblies.
Bashor CJ, Patel N, Choubey S, Beyzavi A, Kondev J, Collins JJ, Khalil AS. Bashor CJ, et al. Science. 2019 May 10;364(6440):593-597. doi: 10.1126/science.aau8287. Epub 2019 Apr 18. Science. 2019. PMID: 31000590 Free PMC article.
References
- Davidson EH. The Regulatory Genome: Gene Regulatory Networks in Development and Evolution. Amsterdam; Boston: Elsevier/Academic; 2006.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous