Histone H3 lysine 79 methyltransferase Dot1 is required for immortalization by MLL oncogenes - PubMed (original) (raw)
Histone H3 lysine 79 methyltransferase Dot1 is required for immortalization by MLL oncogenes
Ming-Jin Chang et al. Cancer Res. 2010.
Abstract
Chimeric oncoproteins resulting from fusion of MLL to a wide variety of partnering proteins cause biologically distinctive and clinically aggressive acute leukemias. However, the mechanism of MLL-mediated leukemic transformation is not fully understood. Dot1, the only known histone H3 lysine 79 (H3K79) methyltransferase, has been shown to interact with multiple MLL fusion partners including AF9, ENL, AF10, and AF17. In this study, we utilize a conditional Dot1l deletion model to investigate the role of Dot1 in hematopoietic progenitor cell immortalization by MLL fusion proteins. Western blot and mass spectrometry show that Dot1-deficient cells are depleted of the global H3K79 methylation mark. We find that loss of Dot1 activity attenuates cell viability and colony formation potential of cells immortalized by MLL oncoproteins but not by the leukemic oncoprotein E2a-Pbx1. Although this effect is most pronounced for MLL-AF9, we find that Dot1 contributes to the viability of cells immortalized by other MLL oncoproteins that are not known to directly recruit Dot1. Cells immortalized by MLL fusions also show increased apoptosis, suggesting the involvement of Dot1 in survival pathways. In summary, our data point to a pivotal requirement for Dot1 in MLL fusion protein-mediated leukemogenesis and implicate Dot1 as a potential therapeutic target.
©2010 AACR.
Conflict of interest statement
Disclosure of Potential Conflicts of Interest
The authors have no conflicts of interest to disclose
Figures
Figure 1
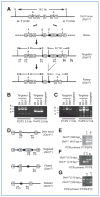
Generation and characterization of Dot1l f/f and Dot1l f/Δ mice. A, targeting vector was cut with _Aat_II to release an 11-kb insert with 4.9-kb 5′ arm (away from the 5′ LoxP site) and 2.6-kb 3′ arm (away from the 3′ FRT sites) for recombination. Boxes with numbers inside indicate exons. The relative positions of _Bam_HI (B), AatII (A) sites, 5′ and 3′ probes used for Southern blot (see Supplementary Fig. S2), and primers for ES clone genotyping, as well as the predicted sizes of _Bam_HI fragments are shown. B and C, PCR-based genotyping of correctly targeted ES cells. Shown are agarose gel analyses of PCR products using the indicated primers. ES cells were microinjected into MF-1 blastocysts for generation of chimeras. D, diagram showing the relative positions of the primers used for mouse genotyping. E–G, PCR-based mouse genotyping. Shown are agarose gel analyses of PCR products from mice carrying different Dot1l alleles.
Figure 2
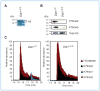
Cre-mediated deletion of Dot1l impairs survival in mouse hematopoietic progenitor cells immortalized by MLL-GAS7, MLL-AFX, and MLL-AF9 but not by E2a-Pbx1. A, experimental scheme to evaluate the effect of Dot1l deletion shows the time points when CFU activity, genotype (by PCR), or Hoxa9 expression (by qRT-PCR) was examined. All experiments were carried out using cells from Dot1l f/f mice unless specified. B, CFUs per 104 cells and representative colony morphologies (20× magnification) of cells immortalized by the indicated fusion oncogenes after Cre-mediated Dot1l deletion. Error bars indicate SD from 3 independent experiments with the exception of E2a-Pbx1, which is from 2 independent experiments. Each independent experiment was conducted in duplicate. Scale bar, 1 mm. C, Wright–Giemsa stain of GFP-sorted cells 6 days after transduction. Representative cells are enlarged in insets to show morphologic details. Magnification is 400 × and scale bars are 40 μm. Arrowheads indicate cells with condensed, fragmented nuclei. D, percentage of GFP+ cells at 3 or 5 days after GFP or Cre-GFP transduction in cells expressing indicated oncogens. Nontransduced cells were used as controls (gray fill) to determine GFP positivity. Cells from _Dot1f/_Δ mice were used in all samples with the exception of the MLL-AFX–immortalized cells, which originated from Dot1lf/f mice. The percentage of GFP+ cells is shown on each histogram. E, Dot1l genomic status was examined by PCR at days 0 and 8 in methylcellulose culture. Arrowhead, floxed allele at 510 bp; open arrowhead, deleted allele at 378 bp. F, relative expression levels of Hoxa9 after Cre-mediated Dot1l deletion. Expression levels are normalized to GAPDH and expressed relative to GFP-transduced cells (set to 100%). Error bars indicate the SD of analyses carried out in triplicate.
Figure 3
Dot1l deletion abolishes histone H3K79 methylation. A, Coomassie-stained SDS-PAGE gel of histone extract with histone H3 band indicated. B, histone H3K79 methylation status in E2a-Pbx1–immortalized cells obtained from Dot1lf/f or Dot1l_Δ/_Δ backgrounds was examined by Western blot using antibodies specific for dimethyl (K79me2) or trimethyl (K79me3) marks. The total histone H3 serves as a loading control. C, SDS-PAGE–purified histone H3 from E2a-Pbx1–immortalized cells with Dot1lf/f or _Dot1lf/_Δ genotype was subjected to LC-MS analysis. The relative abundance of the H3 peptide was manually set to 100. H3 peptide, YRPGTVALR; K79me0, K79me1, and K79me3 represent the unmodified (EIAQDFK), monomethyl and dimethyl (EIAQDFKTDLR) H3K79 peptides, respectively.
Figure 4
Increased Annexin V labeling in Dot1l deleted cells immortalized by MLL-AF9, MLL-GAS7, or MLL-AFX but not by E2a-Pbx1. Immortalized hematopoietic cells expressing the indicated oncogenes were transduced with GFP or Cre-GFP, labeled with Annexin V-PE/7-AAD, and analyzed by flow cytometry 5 days after transduction. The Annexin V-positive/7-AAD-negative cells in the lower right quadrant and the Annexin V–positive/7-AAD-positive cells in the upper right quadrant represent early apoptotic and late apoptotic/necrotic cells, respectively. GFP+ cells are presented as dual parameter contour plots and the percentage of cells in each quadrant is indicated.
Similar articles
- Leukemic transformation by the MLL-AF6 fusion oncogene requires the H3K79 methyltransferase Dot1l.
Deshpande AJ, Chen L, Fazio M, Sinha AU, Bernt KM, Banka D, Dias S, Chang J, Olhava EJ, Daigle SR, Richon VM, Pollock RM, Armstrong SA. Deshpande AJ, et al. Blood. 2013 Mar 28;121(13):2533-41. doi: 10.1182/blood-2012-11-465120. Epub 2013 Jan 29. Blood. 2013. PMID: 23361907 Free PMC article. - MLL-rearranged leukemia is dependent on aberrant H3K79 methylation by DOT1L.
Bernt KM, Zhu N, Sinha AU, Vempati S, Faber J, Krivtsov AV, Feng Z, Punt N, Daigle A, Bullinger L, Pollock RM, Richon VM, Kung AL, Armstrong SA. Bernt KM, et al. Cancer Cell. 2011 Jul 12;20(1):66-78. doi: 10.1016/j.ccr.2011.06.010. Cancer Cell. 2011. PMID: 21741597 Free PMC article. - DOT1L, the H3K79 methyltransferase, is required for MLL-AF9-mediated leukemogenesis.
Nguyen AT, Taranova O, He J, Zhang Y. Nguyen AT, et al. Blood. 2011 Jun 23;117(25):6912-22. doi: 10.1182/blood-2011-02-334359. Epub 2011 Apr 26. Blood. 2011. PMID: 21521783 Free PMC article. - The diverse functions of Dot1 and H3K79 methylation.
Nguyen AT, Zhang Y. Nguyen AT, et al. Genes Dev. 2011 Jul 1;25(13):1345-58. doi: 10.1101/gad.2057811. Genes Dev. 2011. PMID: 21724828 Free PMC article. Review. - Targeting DOT1L and HOX gene expression in MLL-rearranged leukemia and beyond.
Chen CW, Armstrong SA. Chen CW, et al. Exp Hematol. 2015 Aug;43(8):673-84. doi: 10.1016/j.exphem.2015.05.012. Epub 2015 Jun 25. Exp Hematol. 2015. PMID: 26118503 Free PMC article. Review.
Cited by
- Targeted drug discovery for pediatric leukemia.
Napper AD, Watson VG. Napper AD, et al. Front Oncol. 2013 Jul 8;3:170. doi: 10.3389/fonc.2013.00170. eCollection 2013. Front Oncol. 2013. PMID: 23847761 Free PMC article. - An Af9 cis-element directly targets Dot1a to mediate transcriptional repression of the αENaC gene.
Zhang W, Yu Z, Wu H, Chen L, Kong Q, Kone BC. Zhang W, et al. Am J Physiol Renal Physiol. 2013 Feb 15;304(4):F367-75. doi: 10.1152/ajprenal.00537.2011. Epub 2012 Nov 14. Am J Physiol Renal Physiol. 2013. PMID: 23152297 Free PMC article. - Installation of a cancer promoting WNT/SIX1 signaling axis by the oncofusion protein MLL-AF9.
Zhang LS, Kang X, Lu J, Zhang Y, Wu X, Wu G, Zheng J, Tuladhar R, Shi H, Wang Q, Morlock L, Yao H, Huang LJ, Maire P, Kim J, Williams N, Xu J, Chen C, Zhang CC, Lum L. Zhang LS, et al. EBioMedicine. 2019 Jan;39:145-158. doi: 10.1016/j.ebiom.2018.11.039. Epub 2018 Dec 6. EBioMedicine. 2019. PMID: 30528456 Free PMC article. - RNA Polymerase II-Dependent Transcription Initiated by Selectivity Factor 1: A Central Mechanism Used by MLL Fusion Proteins in Leukemic Transformation.
Yokoyama A. Yokoyama A. Front Genet. 2019 Jan 14;9:722. doi: 10.3389/fgene.2018.00722. eCollection 2018. Front Genet. 2019. PMID: 30693017 Free PMC article. Review. - AF10 regulates progressive H3K79 methylation and HOX gene expression in diverse AML subtypes.
Deshpande AJ, Deshpande A, Sinha AU, Chen L, Chang J, Cihan A, Fazio M, Chen CW, Zhu N, Koche R, Dzhekieva L, Ibáñez G, Dias S, Banka D, Krivtsov A, Luo M, Roeder RG, Bradner JE, Bernt KM, Armstrong SA. Deshpande AJ, et al. Cancer Cell. 2014 Dec 8;26(6):896-908. doi: 10.1016/j.ccell.2014.10.009. Epub 2014 Nov 20. Cancer Cell. 2014. PMID: 25464900 Free PMC article.
References
- Lacoste N, Utley RT, Hunter JM, Poirier GG, Côte J. Disruptor of telomeric silencing-1 is a chromatin-specific histone H3 methyltransferase. J Biol Chem. 2002;277:30421–4. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 DK080236/DK/NIDDK NIH HHS/United States
- CA 105049/CA/NCI NIH HHS/United States
- CA 098459/CA/NCI NIH HHS/United States
- R01 CA098459/CA/NCI NIH HHS/United States
- P01 CA105049/CA/NCI NIH HHS/United States
- DK 080236/DK/NIDDK NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases