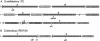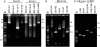Dynamic properties of the Sulfolobus CRISPR/Cas and CRISPR/Cmr systems when challenged with vector-borne viral and plasmid genes and protospacers - PubMed (original) (raw)
Dynamic properties of the Sulfolobus CRISPR/Cas and CRISPR/Cmr systems when challenged with vector-borne viral and plasmid genes and protospacers
Soley Gudbergsdottir et al. Mol Microbiol. 2011 Jan.
Free PMC article
Abstract
The adaptive immune CRISPR/Cas and CRISPR/Cmr systems of the crenarchaeal thermoacidophile Sulfolobus were challenged by a variety of viral and plasmid genes, and protospacers preceded by different dinucleotide motifs. The genes and protospacers were constructed to carry sequences matching individual spacers of CRISPR loci, and a range of mismatches were introduced. Constructs were cloned into vectors carrying pyrE/pyrF genes and transformed into uracil auxotrophic hosts derived from Sulfolobus solfataricus P2 or Sulfolobus islandicus REY15A. Most constructs, including those carrying different protospacer mismatches, yielded few viable transformants. These were shown to carry either partial deletions of CRISPR loci, covering a broad spectrum of sizes and including the matching spacer, or deletions of whole CRISPR/Cas modules. The deletions occurred independently of whether genes or protospacers were transcribed. For family I CRISPR loci, the presence of the protospacer CC motif was shown to be important for the occurrence of deletions. The results are consistent with a low level of random dynamic recombination occurring spontaneously, either inter-genomically or intra-genomically, at the repeat regions of Sulfolobus CRISPR loci. Moreover, the relatively high incidence of single-spacer deletions observed for S. islandicus suggests that an additional more directed mechanism operates in this organism.
© 2010 Blackwell Publishing Ltd.
Figures
Fig. 1
Scheme showing the vector–host constructs. A. pEXA2 was produced by modifying the Sulfolobus_–_E. coli shuttle vector pZC1 (Peng et al., 2009). B. The cloning region of the two vectors showing positions of promoters P1 and P2, where P1 is arabinose-inducible, the terminators T1 to T3, and cloning sites for viral genes and protospacers including a multiple cloning site (MCS) in pEXA2. C. Mutations in the pyrE/pyrF genes of the uracil auxotrophic hosts S. solfataricus LnF1 (Z. Chen et al., unpublished) and S. islandicus E233S (Deng et al., 2009) are indicated.
Fig. 2
Schematic representation of (A) the six CRISPR loci A to F of S. solfataricus P2 and (B) CRISPR loci 1 and 2 of S. islandicus REY15A, drawn to scale (extending from genomic positions 725 000 to 742 500), together with the cassettes of cas genes and cmr genes. CRISPR loci A to D (genomic positions 1 233 400–1 311 600) and CRISPR loci E and F (positions 1 744 000–1 815 500) are clustered within the S. solfataricus P2 genome. The positions of spacers matching protospacers within the vector constructs are indicated where ‘s’ denotes spacer followed by the number of the spacer measured from the leader region, indicated by an arrowhead. Sulfolobales family types of CRISPR/Cas modules are indicated by I and II.
Fig. 3
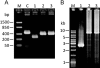
Agarose gels showing examples of PCR products obtained from CRISPR loci of transformants of S. solfataricus P2-InF1 carrying genes for ATV618, ATV145 and ATV892 cloned into pEXA2. A. PCR products obtained from CRISPR loci after challenging with constructs carrying perfect matches to spacers in locus D (ATV618 and ATV145) and locus A (ATV892). The bracketed spacer number denotes the presence of the matching spacer 28. B. PCR products amplified from locus A of transformants with ATV892 carrying two mismatches in the protospacer sequence. C. PCR products from locus D when challenged by ATV618 containing three mismatches in the protospacer, and two samples showing perfect matching protospacers with no transcription. M indicates DNA size markers. The illustrated PCR products yielded the results for estimating the deletion sizes marked with asterisks in Table 1.
Fig. 4
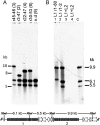
A. Northern blot probing for crRNAs transcribed from spacer 29 of locus D of S. solfataricus P2. The transformant which apparently lacks CRISPR loci A to D (Table 1) was probed. The vector construct carried the ATV618 gene with a matching protospacer and CC motif. Samples were tested without and with arabinose stimulation of ATV618 mRNA transcription to test whether there was any effect of the mRNA. pEXA2 denotes transformants carrying the empty vector with no insert. Normally processed crRNAs are only visible for transformants carrying the empty vector. B. Northern blot probing for the protospacer region of ATV145 transcripts from forward and reverse strands. + denotes the total RNA isolated from the pEXA2-ATV145 transformant and − indicates the total RNA isolated from the transformant of the construct pEXA2-ATV145 for which the araS promoter had been exchanged with an archaeal transcriptional terminator (see main text). ‘P’ represents a positive control where a 10 pmol 22-nt-long DNA oligonucleotide is employed that is complementary to the probe. Sequences of oligonucleotide probes are given in Experimental procedures.
Fig. 5
Agarose gels showing examples of PCR products obtained from CRISPR loci of transformants carrying protospacers and CC and CT PAM motifs cloned into pDL1 and pEXA2 respectively. A. S. islandicus REY15A where both locus 2 carrying the matching spacer and, as a control, locus 1 were amplified. B. S. solfataricus P2. P1 to P7 refer to the experiments listed in Table 3. M indicates DNA size markers. Bracketed spacer numbers denote the presence of the matching spacer. All the illustrated samples carrying deletions are marked with asterisks in Table 3.
Fig. 6
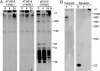
Southern blot analysis of CRISPR locus deletions of S. islandicus REY15A transformants where sample C denotes a control sample of S. islandicus REY15A. The deleted regions determined by sequencing are indicated above each well. A. Analysis of deletions in locus 2 determined from the PCR products shown Fig. 5A where the numbers in the brackets correspond to the sample numbers on agarose gel. B. Samples correspond to the PCR products illustrated in Fig. 7A and B. Tracks 1 and 2 carry transformants 1 and 2 while tracks 3 and 4 carry transformants with protospacers matching both spacer 20 of locus 1 and spacer 45 of locus 2 (Table 5). The lower diagram shows the restriction sites in the CRISPR region where the expected DNA fragment sizes are indicated.
Fig. 7
PCR products showing deletions in CRISPR locus 1 produced by a protospacer matching spacer 1 of locus 1 (Table 5).A. PCR from three transformants using the primers bordering spacer 1 of locus 1 where C denotes a control sample run with S. islandicus REY15A. B. PCR products obtained from the same transformants (1–3) with primers amplifying the whole of locus 1.
Fig. 8
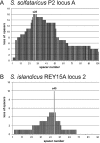
Distributions of deletions within CRISPR loci produced by constructs carrying (A) ATV viral genes or protospacer sequences matching spacer 28 of CRISPR locus A of S. solfataricus P2 and (B) protospacer sequences matching spacer 45 of CRISPR locus 2 of S. islandicus REY15A.
Comment in
- Dangerous weapons: a cautionary tale of CRISPR defence.
Dyall-Smith M. Dyall-Smith M. Mol Microbiol. 2011 Jan;79(1):3-6. doi: 10.1111/j.1365-2958.2010.07451.x. Mol Microbiol. 2011. PMID: 21210528 No abstract available.
Similar articles
- Major and minor crRNA annealing sites facilitate low stringency DNA protospacer binding prior to Type I-A CRISPR-Cas interference in Sulfolobus.
Mousaei M, Deng L, She Q, Garrett RA. Mousaei M, et al. RNA Biol. 2016 Nov;13(11):1166-1173. doi: 10.1080/15476286.2016.1229735. Epub 2016 Sep 12. RNA Biol. 2016. PMID: 27618562 Free PMC article. - Cas4 Nucleases Can Effect Specific Integration of CRISPR Spacers.
Zhang Z, Pan S, Liu T, Li Y, Peng N. Zhang Z, et al. J Bacteriol. 2019 May 22;201(12):e00747-18. doi: 10.1128/JB.00747-18. Print 2019 Jun 15. J Bacteriol. 2019. PMID: 30936372 Free PMC article. - Inter-viral conflicts that exploit host CRISPR immune systems of Sulfolobus.
Erdmann S, Le Moine Bauer S, Garrett RA. Erdmann S, et al. Mol Microbiol. 2014 Mar;91(5):900-17. doi: 10.1111/mmi.12503. Epub 2014 Jan 17. Mol Microbiol. 2014. PMID: 24433295 - SMV1 virus-induced CRISPR spacer acquisition from the conjugative plasmid pMGB1 in Sulfolobus solfataricus P2.
Erdmann S, Shah SA, Garrett RA. Erdmann S, et al. Biochem Soc Trans. 2013 Dec;41(6):1449-58. doi: 10.1042/BST20130196. Biochem Soc Trans. 2013. PMID: 24256236 Free PMC article. Review. - Hot and crispy: CRISPR-Cas systems in the hyperthermophile Sulfolobus solfataricus.
Zhang J, White MF. Zhang J, et al. Biochem Soc Trans. 2013 Dec;41(6):1422-6. doi: 10.1042/BST20130031. Biochem Soc Trans. 2013. PMID: 24256231 Review.
Cited by
- Target motifs affecting natural immunity by a constitutive CRISPR-Cas system in Escherichia coli.
Almendros C, Guzmán NM, Díez-Villaseñor C, García-Martínez J, Mojica FJ. Almendros C, et al. PLoS One. 2012;7(11):e50797. doi: 10.1371/journal.pone.0050797. Epub 2012 Nov 26. PLoS One. 2012. PMID: 23189210 Free PMC article. - CRISPR-Cas Adaptive Immune Systems of the Sulfolobales: Unravelling Their Complexity and Diversity.
Garrett RA, Shah SA, Erdmann S, Liu G, Mousaei M, León-Sobrino C, Peng W, Gudbergsdottir S, Deng L, Vestergaard G, Peng X, She Q. Garrett RA, et al. Life (Basel). 2015 Mar 10;5(1):783-817. doi: 10.3390/life5010783. Life (Basel). 2015. PMID: 25764276 Free PMC article. Review. - Harnessing heterologous and endogenous CRISPR-Cas machineries for efficient markerless genome editing in Clostridium.
Pyne ME, Bruder MR, Moo-Young M, Chung DA, Chou CP. Pyne ME, et al. Sci Rep. 2016 May 9;6:25666. doi: 10.1038/srep25666. Sci Rep. 2016. PMID: 27157668 Free PMC article. - No evidence of inhibition of horizontal gene transfer by CRISPR-Cas on evolutionary timescales.
Gophna U, Kristensen DM, Wolf YI, Popa O, Drevet C, Koonin EV. Gophna U, et al. ISME J. 2015 Sep;9(9):2021-7. doi: 10.1038/ismej.2015.20. Epub 2015 Feb 24. ISME J. 2015. PMID: 25710183 Free PMC article. - Selective Maintenance of Multiple CRISPR Arrays Across Prokaryotes.
Weissman JL, Fagan WF, Johnson PLF. Weissman JL, et al. CRISPR J. 2018 Dec;1(6):405-413. doi: 10.1089/crispr.2018.0034. CRISPR J. 2018. PMID: 31021246 Free PMC article.
References
- Barrangou R, Fremaux C, Deveau H, Richards M, Boyaval P, Moineau S, et al. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315:1709–1712. - PubMed
- Bernander R. The cell cycle of Sulfolobus. Mol Microbiol. 2007;66:557–562. - PubMed
- Bolotin A, Quinquis B, Sorokin A, Ehrlich SD. Clustered regularly interspaced short palindrome repeats (CRISPRs) have spacers of extrachromosomal origin. Microbiology. 2005;151:2551–2561. - PubMed
- Deng L, Zhu H, Chen Z, Liang YX, She Q. Unmarked gene deletion and host–vector system for the hyperthermophilic crenarchaeon Sulfolobus islandicus. Extremophiles. 2009;13:735–746. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources