miR-ID: a novel, circularization-based platform for detection of microRNAs - PubMed (original) (raw)
miR-ID: a novel, circularization-based platform for detection of microRNAs
Pavan Kumar et al. RNA. 2011 Feb.
Abstract
MicroRNAs (miRNAs) are important regulators of gene expression and have great potential as biomarkers, prognostic indicators, and therapeutic targets. Determining the expression patterns of these molecules is essential for elucidating their biogenesis, regulation, relation to disease, and response to therapy. Although PCR-based assays are commonly used for expression profiling of miRNAs, the small size, sequence heterogeneity, and (in some cases) end modifications of miRNAs constrain the performance of existing PCR methods. Here we introduce miR-ID, a novel method that avoids these constraints while providing superior sensitivity and sequence specificity at a lower cost. It also has the unique ability to differentiate unmodified small RNAs from those carrying 2'-OMe groups at their 3'-ends while detecting both forms. miR-ID is comprised of the following steps: (1) circularization of the miRNA by a ligase; (2) reverse transcription of the circularized miRNA (RTC), producing tandem repeats of a DNA sequence complementary to the miRNA; and (3) qPCR amplification of segments of this multimeric cDNA using 5'-overlapping primers and a nonspecific dye such as SYBR Green. No chemically modified probes (e.g., TaqMan) or primers (e.g., LNA) are required. The circular RNA and multimeric cDNA templates provide unmatched flexibility in the positioning of primers, which may include straddling the boundaries between these repetitive miRNA sequences. miR-ID is based on new findings that are themselves of general interest, including reverse transcription of small RNA circles and the use of 5'-overlapping primers for detection of repetitive sequences by qPCR.
Figures
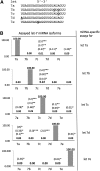
FIGURE 1.
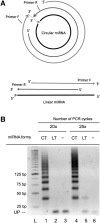
The miR-ID approach. (A) Circularization of a miRNA carrying 5′-phosphate (5′-p) and 3′-hydroxyl (3′-OH) ends by enzymatic ligation (Step 1). (B) Reverse transcription of the circularized miRNA (RTC) by extension of an RT primer, producing a cDNA consisting of tandem repeats of the sequence complementary to the miRNA (Step 2). Examples are shown of RT primers having various alignments relative to the ligation site. (C) qPCR amplification of segments of the multimeric cDNA products of RTC using 5′-overlapping primers together with a nonspecific dye such as SYBR Green (Step 3). (D) Examples of possible alignments of the 5′-overlapping PCR primer pairs with monomer units of the multimeric cDNA, which include straddling the boundaries between these units.
FIGURE 2.
Circularization of miRNAs having nonmethylated 3′-ends (2′-OH miRNAs). (A) Using T4 RNA ligase 1 (Rnl1) at 37°C. (B) Using CircLigase II (CLII) at 60°C. 80 nM synthetic miRNAs (let-7b, let-7g, miR-16, and miR-23a, which have four different nucleotides with 2′-OH and 3′-OH at their 3′-termini), were 32P-labeled at their 5′-ends and then circularized using one or the other ligase. Aliquots were taken at 0, 1, 15, and 60 min of ligation and analyzed on a denaturing 15% polyacrylamide gel. The reaction products include the unmodified linear, 5′-adenylated linear, and circular forms of the miRNAs as shown.
FIGURE 3.
Circularization of miRNAs having a 2′-oxymethyl modification at their 3′-ends (2′-OMe miRNAs). (A) Using T4 RNA ligase 1 (Rnl1) at 37°C. (B) Using CircLigase II (CLII) at 60°C. 80 nM synthetic miRNAs (let-7b, let-7g, miR-16, and miR-23a), which have four different nucleotides with 2′-OMe and 3′-OH at their 3′-termini, were 32P-labeled at their 5′-ends and then circularized using either ligase. Aliquots were taken at 0, 1, 15, and 60 min during the courses of the ligation reactions and analyzed on a denaturing 15% polyacrylamide gel. The reaction products include the unmodified linear, 5′-adenylated linear, and circular forms of the miRNAs as shown.
FIGURE 4.
Reverse transcription of a circular miRNA template (RTC). Circularized synthetic miR-127 miRNA (80 nM) was used as template for SSII reverse transcriptase with 10- and 19-nt RT primers (1 μM) in the presence of [α-32P]dATP. The resulting 32P-labeled primer-extension products were analyzed on a denaturing 12% polyacrylamide gel. (Lane L) A 5′-32P-labeled 10-bp DNA ladder. (Lane 1) Linear 5′-32P-labeled miR-127 as a control. (Lanes 2,3) The extension products of 19- and 10-nt primers, respectively. (Lane 4) A negative control in which the RTC reaction with the 10-nt primer was carried out without the miRNA template.
FIGURE 5.
RT-PCR of circularized miRNA by a pair of 5′-overlapping primers yields multimeric amplicons. (A) Alignment of 5′-overlapping primers with circular (CT) and linear (LT) forms of an miRNA. (B) Synthetic linear and circularized let-7b miRNAs (2 nM) were used as templates for reverse transcription by SSII using Primer R (80 nM), followed by PCR using the same Primer R together with Primer F. Primer R (18 nt) was complementary to nucleotides 1–18 of let-7b miRNA, while Primer F (18 nt) corresponded to nucleotides 4–21 of let-7b. The 5′-ends of Primers F and R have 15 nt of complementarity, leaving 3-nt overhangs at their 3′-ends when hybridized to each other. The PCR products were analyzed on a 3% agarose gel. Lanes 1–3 correspond to 20 cycles, and lanes 4–6 to 25 cycles of PCR. (Lanes 3,6) Negative controls, in which the RT-PCR reactions were carried out without the miRNA templates. (Lane L) A DNA ladder. (UP) Unused PCR primers.
FIGURE 6.
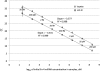
Side-by-side comparison of sensitivity, detection limit, and dynamic range for the miR-ID and TaqMan microRNA assays. Standard curves for both assays were generated for a dilution series from 200 pM to 20 aM of synthetic lin-4 miRNA for either the miR-ID and the TaqMan assays. The data for the TaqMan assay correspond closely to the previously reported standard curve (Chen et al. 2005), taking into account that in the latter publication the concentrations of lin-4 (1.3 aM to 13 pM) represented inputs into the PCR reactions rather than inputs into RT reactions as shown here.
FIGURE 7.
Side-by-side comparison of circularization of 2′-OH miRNA by ligases Rnl1 and CLII. Synthetic let-7b miRNA having 5′-p and 2′-OH/3′-OH ends was used in this example. Standard curves were generated for a dilution series from 200 pM to 20 aM let-7b for miR-ID assays in which either T4 RNA ligase 1 (Rnl1) or CircLigase II (CLII) was used in the circularization step, followed by the usual RTC and qPCR steps.
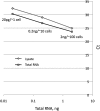
FIGURE 8.
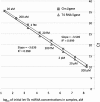
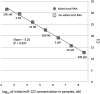
Sensitivity and detection limit of an individual miRNA are not affected by the presence of total cellular RNA. Synthetic miR-127 miRNA having 5′-p and 2′-OH/3′-OH ends was used in this example. Standard curves were generated for a dilution series from 200 pM to 200 aM miR-127 for miR-ID assays performed either in the absence (squares) or the presence of 20 ng of total cellular RNA (diamonds). The total RNA was extracted from Jurkat cells, which do not express miR-127.
FIGURE 9.
Discrimination of let-7 miRNA isoforms. (A) Sequences of selected members of let-7 miRNA family. The nucleotide differences between let-7a and the other isoforms are underlined. (B) Relative detection (in percent) of let-7 isoforms by miR-ID assays, in which every individual isoform was assayed with every isoform-specific primer pair. For each assay, the difference in Ct value for its intended isoform and the other isoforms was converted into a linear percentage, with the fully matched reaction (diagonal values) made equal to 100%. (Bold numbers) miR-ID; (numbers in parentheses) published values (other than 0.0) for QIAGEN's miScript/SYBR Green (*), ABI's TaqMan (**), and LNA/SYBR Green (***) miRNA RT-qPCR assays.
FIGURE 10.
Side-by-side comparison of let-7b detection from purified total RNA versus whole-cell lysates. Standard curves were generated for a dilution series from 20 ng to 20 pg of total RNA (squares) extracted from Jurkat cells or corresponding amounts of lysate (diamonds) of these cells.
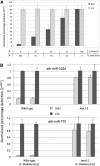
FIGURE 11.
Determination of 2′-_O_-methylation status of miRNAs using miR-ID. (A) Quantification of 2′-OMe and 2′-OH forms of the same miRNA in a mixture. Synthetic let-7b miRNAs having 5′-p and either 2′-OH/3′-OH or 2′-OMe/3′-OH at their 3′-ends were used in this example. These miRNAs were mixed in five predefined proportions, with the total concentration of let-7b RNA constant at 20 pM. Each mixture was subjected to an miR-ID assay in which either Rnl1 or CLII was used in the circularization step, followed by the usual RTC and qPCR steps. For each assay, the Ct value was normalized and converted into a linear percentage, with the CLII assay results made equal to 100%. The Rnl1-based assay gives the concentration of 2′-OH form in the mixture, and normalization to the CLII- based results gives the relative percentage of the 2′-OH and 2′-OMe forms. (B) Identification of plant miRNAs having 2′-OMe or 2′-OH at their 3′-ends. miR-162a and miR-775 miRNAs were assayed in total RNA extracted from wild-type Arabidopsis thaliana plants (Ler) and hen1-1 mutants (Ler). In the wild type, all miRNAs have a 2′-OMe group at the 3′-end, whereas in the mutant, miRNAs are expected to be unmethylated. One hundred nanogram samples of total RNA were subjected to miR-ID assays in which either Rnl1 or CLII was used in the circularization step, followed by the usual RTC and qPCR steps. The Ct values obtained for the CLII-based assays were normalized to 1.00, and the relative signals obtained for Rnl1-based assays were calculated. Two biological replicates for both plant types represented by the gray and black columns are shown.
Similar articles
- MicroRNA detection in prostate tumors by quantitative real-time PCR (qPCR).
Gordanpour A, Nam RK, Sugar L, Bacopulos S, Seth A. Gordanpour A, et al. J Vis Exp. 2012 May 16;(63):e3874. doi: 10.3791/3874. J Vis Exp. 2012. PMID: 22643910 Free PMC article. - Reproducibility of quantitative RT-PCR array in miRNA expression profiling and comparison with microarray analysis.
Chen Y, Gelfond JA, McManus LM, Shireman PK. Chen Y, et al. BMC Genomics. 2009 Aug 28;10:407. doi: 10.1186/1471-2164-10-407. BMC Genomics. 2009. PMID: 19715577 Free PMC article. - High-throughput stem-loop RT-qPCR miRNA expression profiling using minute amounts of input RNA.
Mestdagh P, Feys T, Bernard N, Guenther S, Chen C, Speleman F, Vandesompele J. Mestdagh P, et al. Nucleic Acids Res. 2008 Dec;36(21):e143. doi: 10.1093/nar/gkn725. Epub 2008 Oct 21. Nucleic Acids Res. 2008. PMID: 18940866 Free PMC article. - Expression profiling of microRNA using real-time quantitative PCR, how to use it and what is available.
Benes V, Castoldi M. Benes V, et al. Methods. 2010 Apr;50(4):244-9. doi: 10.1016/j.ymeth.2010.01.026. Epub 2010 Jan 28. Methods. 2010. PMID: 20109550 Review. - Isothermal Amplification for MicroRNA Detection: From the Test Tube to the Cell.
Deng R, Zhang K, Li J. Deng R, et al. Acc Chem Res. 2017 Apr 18;50(4):1059-1068. doi: 10.1021/acs.accounts.7b00040. Epub 2017 Mar 29. Acc Chem Res. 2017. PMID: 28355077 Review.
Cited by
- Right- and left-loop short shRNAs have distinct and unusual mechanisms of gene silencing.
Dallas A, Ilves H, Ge Q, Kumar P, Shorenstein J, Kazakov SA, Cuellar TL, McManus MT, Behlke MA, Johnston BH. Dallas A, et al. Nucleic Acids Res. 2012 Oct;40(18):9255-71. doi: 10.1093/nar/gks662. Epub 2012 Jul 18. Nucleic Acids Res. 2012. PMID: 22810205 Free PMC article. - Decreasing miRNA sequencing bias using a single adapter and circularization approach.
Barberán-Soler S, Vo JM, Hogans RE, Dallas A, Johnston BH, Kazakov SA. Barberán-Soler S, et al. Genome Biol. 2018 Sep 3;19(1):105. doi: 10.1186/s13059-018-1488-z. Genome Biol. 2018. PMID: 30173660 Free PMC article. - Phospho-RNA sequencing with circAID-p-seq.
Del Piano A, Kecman T, Schmid M, Barbieri R, Brocchieri L, Tornaletti S, Firrito C, Minati L, Bernabo P, Signoria I, Lauria F, Gillingwater TH, Viero G, Clamer M. Del Piano A, et al. Nucleic Acids Res. 2022 Feb 28;50(4):e23. doi: 10.1093/nar/gkab1158. Nucleic Acids Res. 2022. PMID: 34850942 Free PMC article. - Specific and sensitive quantitative RT-PCR of miRNAs with DNA primers.
Balcells I, Cirera S, Busk PK. Balcells I, et al. BMC Biotechnol. 2011 Jun 25;11:70. doi: 10.1186/1472-6750-11-70. BMC Biotechnol. 2011. PMID: 21702990 Free PMC article. - Genetically Encoded Reporter Genes for MicroRNA Imaging in Living Cells and Animals.
Song Y, Xu Z, Wang F. Song Y, et al. Mol Ther Nucleic Acids. 2020 Sep 4;21:555-567. doi: 10.1016/j.omtn.2020.06.021. Epub 2020 Jun 27. Mol Ther Nucleic Acids. 2020. PMID: 32721876 Free PMC article. Review.
References
- ABI 2010. Protocol: TaqMan microRNA assays. http://www3.appliedbiosystems.com/cms/groups/mcb_support/documents/gener...
- Ambion 2008. mirVana miRNA Reference Panel v9.1. User information: Real-Time RT-PCR: Using TaqMan microRNA assays; http://tools.invitrogen.com/content/sfs/manuals/sp_4388891.pdf
- Aravin A, Tuschl T 2005. Identification and characterization of small RNAs involved in RNA silencing. FEBS Lett 579: 5830–5840 - PubMed
- Baker M 2010. MicroRNA profiling: separating signal from noise. Nat Methods 7: 687–692 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources