Antibiotics and UV radiation induce competence for natural transformation in Legionella pneumophila - PubMed (original) (raw)
Antibiotics and UV radiation induce competence for natural transformation in Legionella pneumophila
Xavier Charpentier et al. J Bacteriol. 2011 Mar.
Abstract
Natural transformation by competence is a major mechanism of horizontal gene transfer in bacteria. Competence is defined as the genetically programmed physiological state that enables bacteria to actively take up DNA from the environment. The conditions that signal competence development are multiple and elusive, complicating the understanding of its evolutionary significance. We used expression of the competence gene comEA as a reporter of competence development and screened several hundred molecules for their ability to induce competence in the freshwater living pathogen Legionella pneumophila. We found that comEA expression is induced by chronic exposure to genotoxic molecules such as mitomycin C and antibiotics of the fluoroquinolone family. These results indicated that, in L. pneumophila, competence may be a response to genotoxic stress. Sunlight-emitted UV light represents a major source of genotoxic stress in the environment and we found that exposure to UV radiation effectively induces competence development. For the first time, we show that genetic exchanges by natural transformation occur within an UV-stressed population. Genotoxic stress induces the RecA-dependent SOS response in many bacteria. However, genetic and phenotypic evidence suggest that L. pneumophila lacks a prototypic SOS response and competence development in response to genotoxic stress is RecA independent. Our results strengthen the hypothesis that competence may have evolved as a DNA damage response in SOS-deficient bacteria. This parasexual response to DNA damage may have enabled L. pneumophila to acquire and propagate foreign genes, contributing to the emergence of this human pathogen.
Figures
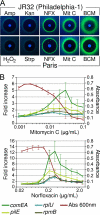
FIG. 1.
Expression of the competence gene comEA is induced by antibiotics and DNA-damaging agents. (A) L. pneumophila strains JR32 (a Philadelphia-1-derived strain) and Paris bearing the comEA-gfp reporter plasmid were grown on CYE plates with paper disks containing selected antibiotics and molecules. All shown molecules produce an inhibition zone (dark area around the disks). Mitomycin C (Mit C), norfloxacin (NFX), and bicyclomycin (BCM) but not hydrogen peroxide, streptomycin (Strp), and ampicillin (Amp) induce expression of comEA-gfp (green halo surrounding the inhibition zone). Photographs were taken with the same exposure time under long-UV light (365 nm). (B) Dose-response induction of comEA-gfp by norfloxacin and mitomycin C. L. pneumophila strain JR32 carrying plasmids with comEA-gfp, pilE-gfp, rplU-gfp, or rpmB-gfp fusions were grown to stationary phase in 96-well plates with increasing concentration of antibiotics. The maximum cell density (_A_600 [Abs 600 nm]) and maximum fold increase in gfp expression relative to cell density (GFP/Abs [_A_600]) are plotted as a function of antibiotic concentration. The maximum cell density displayed on the graphs corresponds to the strain carrying the comEA-gfp fusion. In both graphs, error bars represent standard deviations derived from three independent experiments.
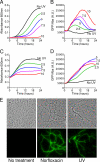
FIG. 2.
UV-irradiated L. pneumophila expresses elevated levels of the competence gene comEA. The JR32 strain bearing the comEA-gfp reporter plasmid was exposed to increasing doses of 254-nm UV radiation (0, 2.5, 5.0, 7.5, and 10 J/m2) and then inoculated into rich medium in microtiter plates at low (_A_600 = 0.02, equivalent to OD600 = 0.1 [A and B]) or high (_A_600 = 0.2, equivalent to OD600 = 0.1 [C and D]) cell densities. Bacterial growth (_A_600 [A and C]) and comEA-gfp expression relative to cell density (GFP/OD, arbitrary units [B and D]) were measured every 10 min over a 24-h period. (E) Microscopic observation of individual L. pneumophila cells after exposure to UV radiation and norfloxacin. The JR32 strain bearing the comEA-gfp reporter plasmid was treated with a single dose of UV radiation (7.5 J/m2) or with a chronic exposure to norfloxacin. After a 24-h growth period, the bacteria were observed using white-light microscopy (top row) and fluorescence microscopy (bottom row). All photographs were taken with the same exposure time.
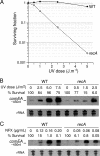
FIG. 3.
RecA is required for survival after UV exposure but is dispensable for the induction of elevated comEA levels after UV irradiation or under chronic exposure to norfloxacin. (A) The L. pneumophila strain Paris and the recA mutant were exposed to increasing doses of UV radiation and inoculated in liquid medium for overnight recovery. The cultures were plated to determine the survival by counting CFU. (B and C) Northern blot analysis of the comEA transcript in wild-type L. pneumophila strain (Paris) and recA mutant after exposure to increasing doses of UV radiation (A) and chronic exposure to increasing doses of norfloxacin (B). The survival of the treated cells is expressed as a percentage of the untreated population. Lower doses of UV radiation and norfloxacin were used with the recA mutant. Ethidium bromide staining of rRNA was used to ensure that equal amounts of total RNA were loaded into each lane.
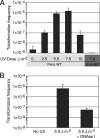
FIG. 4.
Transformability of UV-irradiated L. pneumophila cells. (A) Transformation with exogenous DNA. Wild-type L. pneumophila strain (Paris WT) and transformation-deficient mutant comEC (Δ_comEC_) were exposed to various doses of UV light and inoculated into rich liquid medium in the presence of 1 μg of DNA conferring kanamycin resistance. The cultures were then plated on selective and nonselective media. Transformation frequency represents the number of CFU on selective plates containing kanamycin versus the number of CFU on nonselective plates. (B) Genetic exchange within a stressed population. Two L. pneumophila strains carrying either a gentamicin or a kanamycin chromosomal marker were mixed, exposed to a single dose of UV irradiation (5.0 J/m2), and inoculated in liquid media with or without DNase I (no exogenous DNA added). The transformation frequency represents the number of CFU on selective plates containing kanamycin and gentamicin versus the number of CFU on nonselective plates. In both graphs, error bars represent the standard deviations derived from three independent experiments.
Similar articles
- Induction of competence for natural transformation in Legionella pneumophila and exploitation for mutant construction.
Buchrieser C, Charpentier X. Buchrieser C, et al. Methods Mol Biol. 2013;954:183-95. doi: 10.1007/978-1-62703-161-5_9. Methods Mol Biol. 2013. PMID: 23150395 - SOS response activation and competence development are antagonistic mechanisms in Streptococcus thermophilus.
Boutry C, Delplace B, Clippe A, Fontaine L, Hols P. Boutry C, et al. J Bacteriol. 2013 Feb;195(4):696-707. doi: 10.1128/JB.01605-12. Epub 2012 Nov 30. J Bacteriol. 2013. PMID: 23204467 Free PMC article. - Transposon Insertion Sequencing in a Clinical Isolate of Legionella pneumophila Identifies Essential Genes and Determinants of Natural Transformation.
Hardy L, Juan PA, Coupat-Goutaland B, Charpentier X. Hardy L, et al. J Bacteriol. 2021 Jan 11;203(3):e00548-20. doi: 10.1128/JB.00548-20. Print 2021 Jan 11. J Bacteriol. 2021. PMID: 33168636 Free PMC article. - Speculations on the influence of infecting phenotype on virulence and antibiotic susceptibility of Legionella pneumophila.
Barker J, Brown MR. Barker J, et al. J Antimicrob Chemother. 1995 Jul;36(1):7-21. doi: 10.1093/jac/36.1.7. J Antimicrob Chemother. 1995. PMID: 8537286 Review. - Could DNA uptake be a side effect of bacterial adhesion and twitching motility?
Bakkali M. Bakkali M. Arch Microbiol. 2013 Apr;195(4):279-89. doi: 10.1007/s00203-013-0870-1. Epub 2013 Feb 5. Arch Microbiol. 2013. PMID: 23381940 Free PMC article. Review.
Cited by
- Serum Albumin and Ca2+ Are Natural Competence Inducers in the Human Pathogen Acinetobacter baumannii.
Traglia GM, Quinn B, Schramm ST, Soler-Bistue A, Ramirez MS. Traglia GM, et al. Antimicrob Agents Chemother. 2016 Jul 22;60(8):4920-9. doi: 10.1128/AAC.00529-16. Print 2016 Aug. Antimicrob Agents Chemother. 2016. PMID: 27270286 Free PMC article. - Dissecting the effects of antibiotics on horizontal gene transfer: Analysis suggests a critical role of selection dynamics.
Lopatkin AJ, Sysoeva TA, You L. Lopatkin AJ, et al. Bioessays. 2016 Dec;38(12):1283-1292. doi: 10.1002/bies.201600133. Epub 2016 Oct 4. Bioessays. 2016. PMID: 27699821 Free PMC article. Review. - Amino acids as nutritional factors and (p)ppGpp as an alarmone of the stringent response regulate natural transformation in Micrococcus luteus.
Lichev A, Angelov A, Cucurull I, Liebl W. Lichev A, et al. Sci Rep. 2019 Jul 30;9(1):11030. doi: 10.1038/s41598-019-47423-x. Sci Rep. 2019. PMID: 31363120 Free PMC article. - Subminimal inhibitory concentrations of ampicillin and mechanical stimuli cooperatively promote cell-to-cell plasmid transformation in Escherichia coli.
Kasagaki S, Hashimoto M, Maeda S. Kasagaki S, et al. Curr Res Microb Sci. 2022 Mar 26;3:100130. doi: 10.1016/j.crmicr.2022.100130. eCollection 2022. Curr Res Microb Sci. 2022. PMID: 35909620 Free PMC article. - The cell pole: the site of cross talk between the DNA uptake and genetic recombination machinery.
Kidane D, Ayora S, Sweasy JB, Graumann PL, Alonso JC. Kidane D, et al. Crit Rev Biochem Mol Biol. 2012 Nov-Dec;47(6):531-55. doi: 10.3109/10409238.2012.729562. Epub 2012 Oct 9. Crit Rev Biochem Mol Biol. 2012. PMID: 23046409 Free PMC article. Review.
References
- Almahmoud, I., E. Kay, D. Schneider, and M. Maurin. 2009. Mutational paths toward increased fluoroquinolone resistance in Legionella pneumophila. J. Antimicrob. Chemother. 64:284-293. - PubMed
- Cazalet, C., et al. 2004. Evidence in the Legionella pneumophila genome for exploitation of host cell functions and high genome plasticity. Nat. Genet. 36:1165-1173. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical