Recruitment timing and dynamics of transcription factors at the Hsp70 loci in living cells - PubMed (original) (raw)
Recruitment timing and dynamics of transcription factors at the Hsp70 loci in living cells
Katie L Zobeck et al. Mol Cell. 2010.
Abstract
Chromatin immunoprecipitation (ChIP) studies provide snapshots of factors on chromatin in cell populations. Here, we use live-cell imaging to examine at high temporal resolution the recruitment and dynamics of transcription factors to the inducible Hsp70 loci in individual Drosophila salivary gland nuclei. Recruitment of the master regulator, HSF, is first detected within 20 s of gene activation; the timing of its recruitment resolves from RNA polymerase II and P-TEFb, and these factors resolve from Spt6 and Topo I. Remarkably, the recruitment of each factor is highly synchronous between different cells. In addition, fluorescence recovery after photobleaching (FRAP) analyses show that the entry and exit of multiple factors are progressively constrained upon gene activation, suggesting the gradual formation of a transcription compartment. Furthermore, we demonstrate that poly(ADP-ribose) (PAR) polymerase activity is required to maintain the transcription compartment. We propose that PAR polymers locally retain factors in a transcription compartment.
Copyright © 2010 Elsevier Inc. All rights reserved.
Figures
Figure 1. Colocalization of P-TEFb, Spt6 and Topo I with Pol II at Developmental and Hsp70 Loci in Living Polytene Nuclei
LSCM maximum intensity projections of polytene nuclei co-expressing (A, D) mRFP-P-TEFb (left panel), eGFP-Pol II (middle panel), (B, E) eGFP-Spt6 (left panel) and mRFP-Pol II (middle panel) and (C, F) eGFP-Topo I (left panel) and mRFP-Pol II (middle panel). (A–C) were imaged under NHS conditions and (D–F) are the same nuclei imaged 20 min after HS. Merge presented in right panel. Arrows highlight the position of the Hsp70 loci. Scale bars are 10 μm. See also Figure S1.
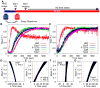
Figure 2. Recruitment Timing of TFs
(A) SDCM was used to obtain 3D images of FP-tagged TFs with the complementary FP-tagged Pol II. First, Drosophila salivary glands were imaged using a room temperature objective, then an objective pre-heated to 36°C was swapped in, causing a nearly instantaneous HS. 3D time series were then obtained continuously over 20 min. (B–C) Normalized TF fluorescence intensities (F.I.) were averaged using 10 sec windows (B) using mean F.I. and (C) using total F.I. HSF in red, Pol II in black, P-TEFb in blue, Spt6 in green, Topo I in purple. Number of samples (n) = 10, 32, 10, 12, and 10, respectively. (D–G) Reproducible differences in recruitment times relative to Pol II were calculated by fitting exponential curves to each set of recruitment data for each factor. (D) HSF (n=10), (E) P-TEFb (n=10), (F) Spt6 (n=12), and (G) Topo I (n=10). Times to reach a specific intensity were calculated and subtracted from the times for Pol II from the same nucleus. Because HSF was not imaged with Pol II, a random set of Pol II curves were used to calculate the paired differences. The red dotted line at Δ _t_=0 represents the time Pol II reaches the corresponding intensity. Error bars represent the SEM. See also Figure S2.
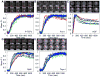
Figure 3. Synchrony in the Recruitment of TFs to Hsp70 loci Among Nuclei of the Same Gland or in Different Glands
(A–E) Representative SDCM recruitment images and corresponding mean F.I. recruitment plots for (A) mRFP-HSF, (B) Pol II (mRFP and eGFP), (C) mRFP-P-TEFb, (D) eGFP-Spt6 and (E) eGFP-Topo I. The 1st image for each factor is the NHS image, 2nd image is the time point before recruitment, while the rest of the images are spaced to depict the recruitment kinetics of the factors (note: HSF is recruited by the first time point after HS). Scale bars are 10 μm. Plots show normalized mean F.I. of factors’ recruitment in nuclei of the same gland (same color lines) and nuclei from different glands (different color lines) over 20 min HS. Each graph represents recruitment data from three or more glands containing 2 or more nuclei from each gland. Red arrows mark the location of the Hsp70 loci. See also Figure S3.
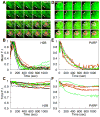
Figure 4. Association of H2B and PARP with Hsp70 Loci after Decondensation
(A and D) Representative time course images illustrating the localization of (A) mRFP-H2B and (D) PARP-eGFP to the Hsp70 loci after HS. Top panel shows the localization of the factor, while the bottom panel shows a merge between the factor (green) and Pol II (red). The 1st image (0 sec) of each panel is the NHS image, 2nd image is the time point before additional Pol II recruitment and consecutive images are spaced out over the course of HS. Arrows indicate the Hsp70 loci, and a progressive decrease in mRFP-H2B or PARP-eGFP intensity can be seen in these images. Scale bars are 10 μm. (B and E) Mean F.I. plots of (B) mRFP-H2B and (E) PARP-eGFP, using Pol II as an indicator for the volume of the Hsp70 loci. Lines of the same color are from the same gland, while different colors are from different glands. 2–3 glands containing 2–4 nuclei each are plotted. (C and F) Total F.I. plots using a constant volume for (C) mRFP-H2B and (F) PARP-eGFP. Lines of the same color are from the same gland, while different colors are from different glands. Fluorescence intensity of 2–4 nuclei each from 2–3 glands are plotted for each factor.
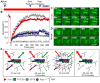
Figure 5. FRAP Dynamics of TFs at the Hsp70 Loci Change After Length of HS
(A, C, and E) FRAP of the three TFs after different lengths of HS: (A) mRFP-P-TEFb, (C) eGFP-Spt6 and (E) eGFP-Topo I. Panels show representative FRAP images. The top panel was bleached after 10 min HS, middle panel was bleached after 20 min HS and lower panel was bleached after 40 min HS (C) or 60 min HS (A and E). All scale bars equal 10 μm. (B, D and F) Plots of Normalized F.I. at the Hsp70 loci. (B) mRFP-P-TEFb, (D) eGFP-Spt6 and (F) eGFP-Topo I. Bleaching resulted in a decrease of 40–60% initial F.I.; however, these plots are normalized to both initial F.I. and bleach depth. Red, 10 min HS; Green, 20 min HS; Blue, 40 min HS; Purple, 60 min HS (n=11, 10, 12, 10 for P-TEFb, respectively; n=13, 6, 6 for Spt6, respectively; n=13, 20, 17, 16 for Topo I, respectively). Error bars represent the SEM. See also Figure S4.
Figure 6. PARP Catalytic Activity is Required for the Maintenance of the Transcription Compartment
(A) Schematic of PJ34 perfusion protocol. Media only or 3μM PJ34 was perfused over the gland for 5 min starting 35 min after HS. FRAP of eGFP-Pol II at the Hsp70 loci was initiated as soon as perfusion stopped. (B–D) FRAP of eGFP-Pol II in PJ34 perfused glands and controls. (B) FRAP curves of eGFP-Pol II perfused with PJ34 (red, n=3) at 40 min HS or with Media only at 40 min HS (blue, n=4) and no perfusion controls at 10 min HS (gray squares, n=4) and 40 min HS (gray circles, n=3). Error bars represent SEM. Representative images of (C) eGFP-Pol II after PJ34 perfusion and (D) after Media only perfusion. Scale bars = 5μm. (E) Model for the progressive formation of transcription compartment during the time course of HS. PARylated proteins accumulate at the Hsp70 loci over HS activation. The accumulation of PAR restricts the ability of proteins to diffuse into and out of the compartment, with individual factors behaving differently, with Spt6 and Pol II unable to exchange with the surrounding nucleoplasm by 40 min HS. Treatment with PJ34 a PARP inhibitor at 40 min HS, reduces the amount of PAR present and restores the ability of Pol II to exchange with the nucleoplasm. Light shaded arrows represent the potential dynamics of the TFs after PJ34 treatment.
Comment in
- The three Rs of transcription: recruit, retain, and recycle.
Motta-Mena LB, Partch CL, Gardner KH. Motta-Mena LB, et al. Mol Cell. 2010 Dec 22;40(6):855-8. doi: 10.1016/j.molcel.2010.12.010. Mol Cell. 2010. PMID: 21172650 Free PMC article.
Similar articles
- Transcription factor and polymerase recruitment, modification, and movement on dhsp70 in vivo in the minutes following heat shock.
Boehm AK, Saunders A, Werner J, Lis JT. Boehm AK, et al. Mol Cell Biol. 2003 Nov;23(21):7628-37. doi: 10.1128/MCB.23.21.7628-7637.2003. Mol Cell Biol. 2003. PMID: 14560008 Free PMC article. - Live-cell imaging of RNA Pol II and elongation factors distinguishes competing mechanisms of transcription regulation.
Versluis P, Graham TGW, Eng V, Ebenezer J, Darzacq X, Zipfel WR, Lis JT. Versluis P, et al. Mol Cell. 2024 Aug 8;84(15):2856-2869.e9. doi: 10.1016/j.molcel.2024.07.009. Mol Cell. 2024. PMID: 39121843 - Dynamics of heat shock factor association with native gene loci in living cells.
Yao J, Munson KM, Webb WW, Lis JT. Yao J, et al. Nature. 2006 Aug 31;442(7106):1050-3. doi: 10.1038/nature05025. Epub 2006 Aug 23. Nature. 2006. PMID: 16929308 - Getting up to speed with transcription elongation by RNA polymerase II.
Jonkers I, Lis JT. Jonkers I, et al. Nat Rev Mol Cell Biol. 2015 Mar;16(3):167-77. doi: 10.1038/nrm3953. Epub 2015 Feb 18. Nat Rev Mol Cell Biol. 2015. PMID: 25693130 Free PMC article. Review. - The heat shock response: A case study of chromatin dynamics in gene regulation.
Teves SS, Henikoff S. Teves SS, et al. Biochem Cell Biol. 2013 Feb;91(1):42-8. doi: 10.1139/bcb-2012-0075. Epub 2013 Feb 13. Biochem Cell Biol. 2013. PMID: 23442140 Review.
Cited by
- Transcription elongation control by the 7SK snRNP complex: Releasing the pause.
McNamara RP, Bacon CW, D'Orso I. McNamara RP, et al. Cell Cycle. 2016 Aug 17;15(16):2115-2123. doi: 10.1080/15384101.2016.1181241. Epub 2016 May 6. Cell Cycle. 2016. PMID: 27152730 Free PMC article. Review. - Temporal Dissection of Rate Limiting Transcriptional Events Using Pol II ChIP and RNA Analysis of Adrenergic Stress Gene Activation.
Morris DP, Lei B, Longo LD, Bomsztyk K, Schwinn DA, Michelotti GA. Morris DP, et al. PLoS One. 2015 Aug 5;10(8):e0134442. doi: 10.1371/journal.pone.0134442. eCollection 2015. PLoS One. 2015. PMID: 26244980 Free PMC article. - A transcriptional cycling model recapitulates chromatin-dependent features of noisy inducible transcription.
Bullock ME, Moreno-Martinez N, Miller-Jensen K. Bullock ME, et al. PLoS Comput Biol. 2022 Sep 9;18(9):e1010152. doi: 10.1371/journal.pcbi.1010152. eCollection 2022 Sep. PLoS Comput Biol. 2022. PMID: 36084132 Free PMC article. - Controlling gene expression in response to stress.
de Nadal E, Ammerer G, Posas F. de Nadal E, et al. Nat Rev Genet. 2011 Nov 3;12(12):833-45. doi: 10.1038/nrg3055. Nat Rev Genet. 2011. PMID: 22048664 Review. - Biochemical Timekeeping Via Reentrant Phase Transitions.
Portz B, Shorter J. Portz B, et al. J Mol Biol. 2021 Jun 11;433(12):166794. doi: 10.1016/j.jmb.2020.166794. Epub 2020 Dec 31. J Mol Biol. 2021. PMID: 33387533 Free PMC article. Review.
References
- Althaus FR, Hofferer L, Kleczkowska HE, Malanga M, Naegeli H, Panzeter PL, Realini CA. Histone shuttling by poly ADP-ribosylation Mol. Cell Biochem. 1994;138:53–59. - PubMed
- Beermann W. Chromomeres and genes. Results Probl Cell Differ. 1972;4:1–33. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- F32 GM087003/GM/NIGMS NIH HHS/United States
- GM25232/GM/NIGMS NIH HHS/United States
- R37 GM025232/GM/NIGMS NIH HHS/United States
- R01 GM025232/GM/NIGMS NIH HHS/United States
- R01 GM025232-33/GM/NIGMS NIH HHS/United States
- R01 GM025232-32/GM/NIGMS NIH HHS/United States
- GM087003/GM/NIGMS NIH HHS/United States
- R01 GM025232-34/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous