Computational and experimental identification of mirtrons in Drosophila melanogaster and Caenorhabditis elegans - PubMed (original) (raw)
Computational and experimental identification of mirtrons in Drosophila melanogaster and Caenorhabditis elegans
Wei-Jen Chung et al. Genome Res. 2011 Feb.
Abstract
Mirtrons are intronic hairpin substrates of the dicing machinery that generate functional microRNAs. In this study, we describe experimental assays that defined the essential requirements for entry of introns into the mirtron pathway. These data informed a bioinformatic screen that effectively identified functional mirtrons from the Drosophila melanogaster transcriptome. These included 17 known and six confident novel mirtrons among the top 51 candidates, and additional candidates had limited read evidence in available small RNA data. Our computational model also proved effective on Caenorhabditis elegans, for which the identification of 14 cloned mirtrons among the top 22 candidates more than tripled the number of validated mirtrons in this species. A few low-scoring introns generated mirtron-like read patterns from atypical RNA structures, but their paucity suggests that relatively few such loci were not captured by our model. Unexpectedly, we uncovered examples of clustered mirtrons in both fly and worm genomes, including a <8-kb region in C. elegans harboring eight distinct mirtrons. Altogether, we demonstrate that discovery of functional mirtrons, unlike canonical miRNAs, is amenable to computational methods independent of evolutionary constraint.
Figures
Figure 1.
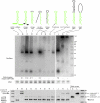
Constructs used for structural analysis of mirtron biogenesis. Shown are sequence variants of the mir-1003 mirtron used for functional tests. (Green) The mature miRNA sequence; (yellow) the nucleotides differing from mir-1003. Their relative abilities to be processed in S2 cells are indicated (see also Fig. 2).
Figure 2.
Structure-function analysis of mirtron biogenesis. (Top) S2 cells were transfected with UAS-mirtron and ub-Gal4 plasmids and RNA was isolated and subjected to Northern blot using an LNA probe antisense to miR-1003. Ethidium bromide staining of 5S rRNA is shown as a loading control. The fold increase in mature miR-1003 above control transfections is indicated below; (−) No substantial increase in miR-1003 level (>2 folds) was detected. (A) Control transfection using empty expression vector shows that S2 cells express a low level of the mirtron-derived miRNA miR-1003. (B) Introduction of mir-1003 expression plasmid, which includes portions of its endogenous flanking exons, yields strongly elevated pre-mir-1003 and mature miR-1003. Neither substitution of mir-1003 exonic context (C), nor replacement of its terminal loop (D), interferes with its biogenesis. Extensive mutation of its miRNA* arm abolishes production of miR-1003 (E,G), although a small amount of pre-miRNA is detected in the later case. However, extensive mutation while maintaining hairpin structure supports efficient mirtron biogenesis (F). (H) Introduction of a 5′ hairpin overhang abolishes small RNA production. (I) Extension of the 3′ hairpin overhang strongly impairs mirtron processing, although pre-miRNA accumulated. (J–L) Starting with a terminal loop mutant of mir-1003 (J, see also lane D), structured (K), and unstructured (L) hairpin extensions were introduced. Both constructs yielded substantial amounts of ∼150 nt pre-miRNA product, with higher levels of the fully duplexed intron (K); however, neither supported accumulation of mature miRNA. A ∼75-nt product corresponding to approximately half of the long hairpin intron accumulated; its biogenesis is not known. (Bottom) The same RNA samples used for Northern blotting at top were subjected to RT-PCR analysis to verify splicing accuracy of the mirtron variants. We observed weaker bands for the unspliced products and stronger bands for the spliced products; the DNA template controls at the right provide a size marker to gauge the unspliced amplification products. Note that the wild-type mir-1003 construct in its native CG6995 context includes more exon sequence than the other constructs, leading to the larger sizes of its RT-PCR products (“B”).
Figure 3.
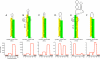
Examples of known and novel mirtrons in D. melanogaster. The abundant small RNAs derived from each hairpin are highlighted, green for the miRNA and yellow for the miRNA*. Below the secondary structures are plots that show the abundance of cloned small RNAs across the aggregate D. melanogaster small RNA data. The small RNA density is highest at either end of each intron, with typically one side accumulating to a higher level; often this is the 3′ arm, but occasionally it is the 5′ arm. The black boxes below the graph indicate the exon–intron boundaries. (A) CG6695_in5/mir-1003 is an example of a conserved, abundantly expressed mirtron with optimal features, including a straight short intronic hairpin with a 2-nt 3′ overhang. (B) Vha-SFD_in3/mir-1006 is an example of a conserved, abundant-expressed mirtron with a large asymmetric internal loop (5 + 2 nt). CG1941_in5 (C) and Cyp4aa1_in3 (D) are novel mirtrons with typical straight hairpins and compatible overhangs. (E) RhoGAP1A_in3 is an expressed mirtron with an unusually large, unstructured terminal loop, a large asymmetric internal loop (5 + 3 nt) and single nucleotide overhangs at its 5′ and 3′ ends. (F) CG15539_in3 exhibits convincing mirtron features, but is on the borderline of confident cloning evidence; nevertheless, its reads exhibit a characteristic 2-nt 3′ overhang on the Dicer-1-cleaved end.
Figure 4.
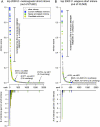
Performance of the computational model for mirtron identification on the D. melanogaster and C. elegans genomes. (A) Performance of an SVM trained on the 14 original D. melanogaster mirtrons (mir-1003-mir-1016) and run across the fly genome. (B) Performance of the D. melanogaster model on C. elegans. In both cases we used as input the annotated short introns 50–120 nt in length; no evolutionary features were considered. The top graphs plot the scores of mirtron likelihood and illustrate that the scores quickly drop following the top predicted candidates. Highlighted in blue are mirtrons previously deposited in miRBase (note that the previously annotated C. elegans mir-2220 was reported earlier but not recognized as a mirtron; it is nonetheless included in the “blue” loci), novel mirtrons annotated in this study are in green, and candidate mirtrons are highlighted in gold. The bottom graphs utilize the same _x_-axis and plot the numbers of validated and candiate mirtrons in consecutive bins of 20 introns in the rank order. Note that a few validated mirtrons scored poorly, and most of these have atypical 3′ overhangs. The full rankings can be viewed in Supplemental Tables S3 and S4.
Figure 5.
CG17560_in3 generates a mirtron from an alternatively spliced intron. Shown is a multiple sequence alignment and phastCons assessment of conservation (obtained from the UCSC Genome Browser). The splice acceptor used to generate the protein-coding transcript is highly conserved across the 12 sequenced Drosophilids; a different splice acceptor is used to generate the CG17560 mirtron. Small RNA mappings exhibit typical Dicer-1 cleavage patterns, including the generation of rare reads corresponding to the cleaved terminal loop. Other rare reads were not summarized in this schematic. Note the slightly atypical hairpin end of this mirtron, which terminates in a 3-nt 3′overhang. Usage of the mirtronic splice generates a frame-shift, since the typical splice site joins in the +2 coding frame, while the mirtron-spliced site joins in the +1 coding frame.
Figure 6.
Exceptional fly and worm mirtrons exhibit strongly unpaired hairpin termini. It is generally accepted that a defined short 3′ overhang is critical for nuclear export of pre-miRNA hairpins via exportin 5. Consequently, a strongly unpaired hairpin base is unfavorable for pre-miRNA maturation. (A) The exceptional C. elegans mirtron mir-1019 exhibits a 2 + 5 hairpin overhang, but still exhibits a typical pattern of mirtronic reads corresponding specifically to the ends of the intron. (B) Similarly, the atypical D. melanogaster mirtron CG3225_in2 exhibits strong evidence for Dicer-1 cleavage despite a 4 + 7 hairpin overhang, including a rare read corresponding to the cleaved terminal loop (highlighted in blue). Reads from this intron exhibit evidence for loading to the siRNA effector AGO2 instead of the miRNA effector, AGO1. Head data including AGO1-IP and oxidized RNA (which enriches for mature AGO2-loaded siRNAs) were reported by Ghildiyal et al. (2010) and S2 cell data from AGO1-IP and AGO2-IP were reported by Czech et al. (2008); to permit comparison between the total and IP levels, these read numbers were normalized per million mapped reads in each library. Note that these worm and fly mirtrons are further atypical in that their mature cloned species derive from their 5p arms; this correlates with the strong thermodynamic asymmetry associated with their unpaired hairpin bases. These mirtrons are exceptional, and few other introns with similarly unpaired bases were productively converted into short cloned RNAs.
Figure 7.
Clustered mirtrons in the D. melanogaster and C. elegans genomes. (A) Drosophila CG1718 generates mirtrons from both its second and third introns; CG1718_in2 was newly identified in this study. Curiously, while the hairpin structure of CG1718_in2 is seemingly suboptimal compared with the previously identified mir-1007, mature miRNAs accumulate to relatively similar levels from these mirtrons. Analysis of head libraries published by Ghildiyal et al. (2010) provided evidence that these mirtrons are expressed in the head and generate RNAs that populate AGO1, but not AGO2 complexes; this study used oxidation (oxi) of input samples to enrich for 2'O-methylated RNAs in mature AGO2 complexes. To permit comparison between the total and IP levels, these read numbers were normalized per million mapped reads in each library. Rarer reads were not shown, except for the informative cloned terminal loops that report on endogenous Dicer-1 processing; the full read patterns are available at
http://cbio.mskcc.org/leslielab/mirtrons
. (B) NM_071513 and NM_071540 are related genes that reside ∼70 kb apart on C. elegans chromosome V. Each gene bears a mirtron whose 3p arm is identical; thus, small RNA reads from this arm map to both mirtrons. We normalized the read numbers to assign half to each locus. On the basis of unique star arms, we can definitively annotate the expression of NM_071540. However, given that the hairpin of NM_071513 has only small symmetric loops, we infer that its processing should be equivalent, if not more efficient, to its paralog. (C) A supercluster of mirtron genes on C. elegans chromosome X. This <8-kb region was previously annotated to contain mir-1018 and mir-2220, of which mir-1018 was previously noted to be a mirtron (Ruby et al. 2007a). Although mir-2220 was earlier annotated as a canonical miRNA (Kato et al. 2009), we infer that it is similarly a mirtron, as its cloned RNAs begin and end with effective splice junctions. Here, we identify six additional mirtrons in this genomic region. Of these, NM_075943_in1 might appear to be a tailed mirtron based on the annotated splice junction; however, that its abundant 3p reads end with CAG suggests that it may be the product of alternative splicing, as seen for the Drosophila mirtron CG17560. Note that in all gene alignments only a subset of informative singleton reads, typically belonging to mirtron star species are shown.
Similar articles
- Analysis of Nearly One Thousand Mammalian Mirtrons Reveals Novel Features of Dicer Substrates.
Wen J, Ladewig E, Shenker S, Mohammed J, Lai EC. Wen J, et al. PLoS Comput Biol. 2015 Sep 1;11(9):e1004441. doi: 10.1371/journal.pcbi.1004441. eCollection 2015 Sep. PLoS Comput Biol. 2015. PMID: 26325366 Free PMC article. - Mammalian mirtron genes.
Berezikov E, Chung WJ, Willis J, Cuppen E, Lai EC. Berezikov E, et al. Mol Cell. 2007 Oct 26;28(2):328-36. doi: 10.1016/j.molcel.2007.09.028. Mol Cell. 2007. PMID: 17964270 Free PMC article. - Intronic microRNA precursors that bypass Drosha processing.
Ruby JG, Jan CH, Bartel DP. Ruby JG, et al. Nature. 2007 Jul 5;448(7149):83-6. doi: 10.1038/nature05983. Epub 2007 Jun 24. Nature. 2007. PMID: 17589500 Free PMC article. - Mirtrons: microRNA biogenesis via splicing.
Westholm JO, Lai EC. Westholm JO, et al. Biochimie. 2011 Nov;93(11):1897-904. doi: 10.1016/j.biochi.2011.06.017. Epub 2011 Jun 21. Biochimie. 2011. PMID: 21712066 Free PMC article. Review. - The long and short of inverted repeat genes in animals: microRNAs, mirtrons and hairpin RNAs.
Okamura K, Chung WJ, Lai EC. Okamura K, et al. Cell Cycle. 2008 Sep 15;7(18):2840-5. doi: 10.4161/cc.7.18.6734. Epub 2008 Sep 5. Cell Cycle. 2008. PMID: 18769156 Free PMC article. Review.
Cited by
- Recent Molecular Genetic Explorations of Caenorhabditis elegans MicroRNAs.
Ambros V, Ruvkun G. Ambros V, et al. Genetics. 2018 Jul;209(3):651-673. doi: 10.1534/genetics.118.300291. Genetics. 2018. PMID: 29967059 Free PMC article. - Analysis of Nearly One Thousand Mammalian Mirtrons Reveals Novel Features of Dicer Substrates.
Wen J, Ladewig E, Shenker S, Mohammed J, Lai EC. Wen J, et al. PLoS Comput Biol. 2015 Sep 1;11(9):e1004441. doi: 10.1371/journal.pcbi.1004441. eCollection 2015 Sep. PLoS Comput Biol. 2015. PMID: 26325366 Free PMC article. - Discovery of hundreds of mirtrons in mouse and human small RNA data.
Ladewig E, Okamura K, Flynt AS, Westholm JO, Lai EC. Ladewig E, et al. Genome Res. 2012 Sep;22(9):1634-45. doi: 10.1101/gr.133553.111. Genome Res. 2012. PMID: 22955976 Free PMC article. - Selective Suppression of the Splicing-Mediated MicroRNA Pathway by the Terminal Uridyltransferase Tailor.
Bortolamiol-Becet D, Hu F, Jee D, Wen J, Okamura K, Lin CJ, Ameres SL, Lai EC. Bortolamiol-Becet D, et al. Mol Cell. 2015 Jul 16;59(2):217-28. doi: 10.1016/j.molcel.2015.05.034. Epub 2015 Jul 2. Mol Cell. 2015. PMID: 26145174 Free PMC article. - Deep annotation of Drosophila melanogaster microRNAs yields insights into their processing, modification, and emergence.
Berezikov E, Robine N, Samsonova A, Westholm JO, Naqvi A, Hung JH, Okamura K, Dai Q, Bortolamiol-Becet D, Martin R, Zhao Y, Zamore PD, Hannon GJ, Marra MA, Weng Z, Perrimon N, Lai EC. Berezikov E, et al. Genome Res. 2011 Feb;21(2):203-15. doi: 10.1101/gr.116657.110. Epub 2010 Dec 22. Genome Res. 2011. PMID: 21177969 Free PMC article.
References
- Batuwita R, Palade V 2009. microPred: Effective classification of pre-miRNAs for human miRNA gene prediction. Bioinformatics 25: 989–995 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 GM083300/GM/NIGMS NIH HHS/United States
- U01 HG004261/HG/NHGRI NIH HHS/United States
- R01-GM083300/GM/NIGMS NIH HHS/United States
- U01-HG004261/HG/NHGRI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases