Perturbation of host nuclear membrane component RanBP2 impairs the nuclear import of human immunodeficiency virus -1 preintegration complex (DNA) - PubMed (original) (raw)
Perturbation of host nuclear membrane component RanBP2 impairs the nuclear import of human immunodeficiency virus -1 preintegration complex (DNA)
Ruonan Zhang et al. PLoS One. 2010.
Abstract
HIV-1 is a RNA virus that requires an intermediate DNA phase via reverse transcription (RT) step in order to establish productive infection in the host cell. The nascent viral DNA synthesized via RT step and the preformed viral proteins are assembled into pre-integration complex (PIC) in the cell cytoplasm. To integrate the viral DNA into the host genome, the PIC must cross cell nuclear membrane through the nuclear pore complex (NPC). RanBP2, also known as Nup358, is a major component of the cytoplasmic filaments that emanates from the nuclear pore complex and has been implicated in various nucleo-cytoplasmic transport pathways including those for HIV Rev-protein. We sought to investigate the role of RanBP2 in HIV-1 replication. In our investigations, we found that RanBP2 depletion via RNAi resulted in profound inhibition of HIV-1 infection and played a pivotal role in the nuclear entry of HIV DNA. More precisely, there was a profound decline in 2-LTR DNA copies (marker for nuclear entry of HIV DNA) and an unchanged level of viral reverse transcription in RanBP2-ablated HIV-infected cells compared to RanBP3-depleted or non-specific siRNA controls. We further demonstrated that the function of Rev was unaffected in RanBP2-depleted latently HIV infected cells (reactivated). We also serendipitously found that RanBP2 depletion inhibited the global ectopic gene expression. In conclusion, RanBP2 is a host factor that is involved in the nuclear import of HIV-1 PIC (DNA), but is not critical to the nuclear export of the viral mRNAs or nucleo-cytoplasmic shuttling of Rev. RanBP2 could be a potential target for efficient inhibition of HIV.
Conflict of interest statement
Competing Interests: The authors have declared that no competing interests exist.
Figures
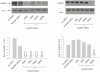
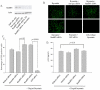
Figure 1. siRNA-mediated depletion of RanBP2 and RanBP3.
SVGA cells were transfected either with RanBP2 siRNA or RanBP3 siRNA, respectively. In parallel, control SVGA cells were either transfected with 200 nM scrambled siRNA or without siRNA. Total proteins were extracted 72 h post siRNA transfection. Western blot analysis was performed to examine levels of RanBP2 and RanBP3, respectively. Actin was used as internal control. Image J was used to analyze the intensities of the bands.
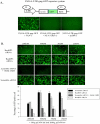
Figure 2. RanBP2 depletion impairs biological activities of ectopically expressed Tat and Rev HIV proteins.
(A) Schematic diagram of Tat/Rev responsive SVGA-LTR-gag-GFP cells is shown. The cells were transfected with either 50 ng NL4-3 plasmid, or 50 ng NL4-3 (Rev-). Fluorescent images were captured 48 h post transfection. (B) SVGA-LTR-gag-GFP reporter cells were transfected either with RanBP2 siRNA, RanBP3 siRNA (negative control) or scrambled siRNA (control). 48 h later, the cells were then co-transfected with 50 ng pCMV-Tat and 200 ng pCMV-Rev. In parallel, Leptomycin B was used as positive control and was added 4 h after the transfection of reporter cells (pre-transfected with scrambled siRNA) with pCMV-Tat and pCMV-Rev. The images were captured and GFP positive cells were analyzed by flow cytometry 48 h post plasmid transfection.
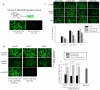
Figure 3. RanBP2 depletion inhibits global ectopic gene expression.
(A) Schematic diagram of Tat responsive SVGA-LTR-GFP reporter cells is shown. Cells were either transfected with 500 ng pCMV-Tat or pcDNA vector (control) and images were captured 48 h after transfection. (B) SVGA-LTR-GFP reporter cells were transfected either with series of concentrations of RanBP2 siRNA or scrambled siRNA. 48 h later, the cells were transfected with 500 ng pCMV-Tat. After 48 h the images were captured and GFP positive cells were analyzed by flow cytometry. (C) SVGA cells were transfected either with 100 nM RanBP2 siRNA, 200 nM RanBP3 siRNA or 100 nM scrambled siRNA. 48 h later, the cells were then transfected either with 100 ng pEGFP, 100 ng pTat-GFP or 100 ng pΔTat-GFP plasmids respectively. The GFP expression was examined 48 h later by capturing the images and analysis by flow cytometry.
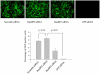
Figure 4. RanBP2 depletion does not inhibit GFP expression in SVGA-GFP transgenic cells.
Stable SVGA-GFP cells were transfected overnight either with 100 nM RanBP2 siRNA, 200 nM RanBP3 siRNA (negative control), 100 nM scrambled siRNA (control), or 100 nM GFP siRNA as a positive control. The cells were then followed up for 5 days. On day 3 images were captured and the GFP positive cells were analyzed by flow cytometry.
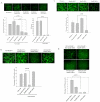
Figure 5. RanBP2 depletion does not interfere with nuclear export of viral mRNA species.
(A) Latently HIV infected monocytic (THP89) cells were transfected with 100 nM RanBP2 siRNA and total proteins were extracted 48 or 72 h post transfection. Western blot analysis revealed the levels of RanBP2 protein. RanBP2 level in cells that were transfected with 100 nM RanBP2 siRNA followed by bryostatin treatment were also examined the performance on viral activity. THP89 cells either transfected with scramble siRNA or left untransfected were used controls. (B–D) THP89 cells were first transfected either with RanBP2 siRNA, RanBP3 siRNA, scrambled siRNA or GFP siRNA as a positive control. 48 h later, the cells were reactivated with 25 ng/ml bryostatin (potent reactivator of latent HIV infection). The GFP expression was examined 48 hrs after bryostatin treatment. (B) The Images were captured and (C) GFP positive cells were analyzed by flow cytometry. (D) Supernatants collected in (B) were analyzed for p24 levels by ELISA.
Figure 6. RanBP2 depletion inhibits HIV-1 replication and lentiviral vector gene expression.
(A) Magi cells were transfected either with RanBP2 siRNA, RanBP3 siRNA (negative control), TNPO3 siRNA (positive control) or scrambled siRNA (control). 48 h later, the cells were transduced with 200 ng/ml p24 equivalent VSV-NLENY1 (recombinant HIV virus containing YFP) for 16 h. The fluorescent images were taken 48 h post transduction followed by collection of the culture supernatants for p24 (ELISA). The YFP positive cells were analyzed by flow cytometry (p<0.01). (B) SVGA cells were transfected either with RanBP2 siRNA, RanBP3 siRNA or scrambled siRNA. 48 h later, the cells were transduced with 200 ng/ml p24 equivalent VSV-NLENY1 for 16 h. The fluorescent images were captured 48 h post transduction by digital camera (Nikon) and YFP positive cells were analyzed by flow cytometry (p<0.001). (C) SVGA-LTR-GFP reporter cells were transduced either with RanBP2 siRNA, RanBP3 siRNA (negative control), scramble siRNA or TNPO3 siRNA as a positive control. 48 hrs later, the cells were then transduced with 200 ng/ml p24 equivalent VSV-NL4-3. Images were captured 48 h after transduction and the GFP positive cells were analyzed by flow cytometry (p<0.01). (D) SVGA cells were transfected either with RanBP2 siRNA, RanBP3 siRNA, TNPO3 siRNA or scrambled siRNA. 48 h later, the cells were transduced with VSV-G pseudotyped lentiviral vector. The fluorescent images were captured 48 h after transduction and GFP positive cells were analyzed by flow cytometry (p<0.01).
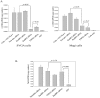
Figure 7. RanBP2 depletion inhibits 2-LTR circle formation (HIV DNA) but not viral reverse transcription.
SVGA and Magi cells were transfected either with RanBP2 siRNA, RanBP3 siRNA (negative control), scrambled siRNA or TNPO3 siRNA as a positive control. 48 h later, the cells were transduced with 200 ng/ml p24 equivalent VSV-NL4-3 (HIV). AZT, a reverse transcriptase inhibitor, was used as a positive control to inhibit the reverse transcription and added 30 min before VSV-HIV transduction. (A) Total DNA was extracted 24 h after VSV-HIV transduction and 2-LTR levels were examined by real-time PCR. (B) Total DNA from SVGA cells was extracted 12 hrs after VSV-HIV transduction and the viral reverse transcription was examined by real-time PCR.
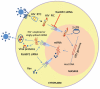
Figure 8. Schematic diagram illustrating role of RanBP2 in HIV-1 DNA (PIC) nuclear import.
HIV-1 entry follows partial uncoating of HIV particles with concomitant viral RNA reverse transcription into cDNA and assembly into PIC with viral proteins MA, Vpr and integrase. The PIC import into the nucleus is facilitated by nuclear membrane component RanBP2. RanBP2 depletion via RNAi inhibits PIC import into the nucleus. The PIC import is detected by the 2-LTR circles (marker of HIV DNA nuclear import). RanBP2 is not involved in HIV-Rev protein nuclear import/export as demonstrated by HIV replication (GFP expression as a marker or viral p24 antigen) seen in HIV latent THP89 model (HIV reactivation by bryostatin).
Similar articles
- The nuclear pore component Nup358 promotes transportin-dependent nuclear import.
Hutten S, Wälde S, Spillner C, Hauber J, Kehlenbach RH. Hutten S, et al. J Cell Sci. 2009 Apr 15;122(Pt 8):1100-10. doi: 10.1242/jcs.040154. Epub 2009 Mar 19. J Cell Sci. 2009. PMID: 19299463 - The nucleoporin Nup358/RanBP2 promotes nuclear import in a cargo- and transport receptor-specific manner.
Wälde S, Thakar K, Hutten S, Spillner C, Nath A, Rothbauer U, Wiemann S, Kehlenbach RH. Wälde S, et al. Traffic. 2012 Feb;13(2):218-33. doi: 10.1111/j.1600-0854.2011.01302.x. Epub 2011 Nov 21. Traffic. 2012. PMID: 21995724 - Human nucleoporins promote HIV-1 docking at the nuclear pore, nuclear import and integration.
Di Nunzio F, Danckaert A, Fricke T, Perez P, Fernandez J, Perret E, Roux P, Shorte S, Charneau P, Diaz-Griffero F, Arhel NJ. Di Nunzio F, et al. PLoS One. 2012;7(9):e46037. doi: 10.1371/journal.pone.0046037. Epub 2012 Sep 25. PLoS One. 2012. PMID: 23049930 Free PMC article. - The HIV-1 Capsid: From Structural Component to Key Factor for Host Nuclear Invasion.
Scoca V, Di Nunzio F. Scoca V, et al. Viruses. 2021 Feb 10;13(2):273. doi: 10.3390/v13020273. Viruses. 2021. PMID: 33578999 Free PMC article. Review. - [HIV-1 Rev and related inhibitors].
Cao Y, Liu XY. Cao Y, et al. Yao Xue Xue Bao. 2007 Apr;42(4):347-51. Yao Xue Xue Bao. 2007. PMID: 17633198 Review. Chinese.
Cited by
- MxB impedes the NUP358-mediated HIV-1 pre-integration complex nuclear import and viral replication cooperatively with CPSF6.
Xie L, Chen L, Zhong C, Yu T, Ju Z, Wang M, Xiong H, Zeng Y, Wang J, Hu H, Hou W, Feng Y. Xie L, et al. Retrovirology. 2020 Jun 29;17(1):16. doi: 10.1186/s12977-020-00524-2. Retrovirology. 2020. PMID: 32600399 Free PMC article. - Capsid-host interactions for HIV-1 ingress.
Jang S, Engelman AN. Jang S, et al. Microbiol Mol Biol Rev. 2023 Dec 20;87(4):e0004822. doi: 10.1128/mmbr.00048-22. Epub 2023 Sep 26. Microbiol Mol Biol Rev. 2023. PMID: 37750702 Free PMC article. Review. - HIV-1 Capsid Core: A Bullet to the Heart of the Target Cell.
Toccafondi E, Lener D, Negroni M. Toccafondi E, et al. Front Microbiol. 2021 Apr 1;12:652486. doi: 10.3389/fmicb.2021.652486. eCollection 2021. Front Microbiol. 2021. PMID: 33868211 Free PMC article. Review. - Cellular cofactors of lentiviral integrase: from target validation to drug discovery.
Taltynov O, Desimmie BA, Demeulemeester J, Christ F, Debyser Z. Taltynov O, et al. Mol Biol Int. 2012;2012:863405. doi: 10.1155/2012/863405. Epub 2012 Aug 7. Mol Biol Int. 2012. PMID: 22928108 Free PMC article. - Modeling HIV-1 nuclear entry with nucleoporin-gated DNA-origami channels.
Shen Q, Feng Q, Wu C, Xiong Q, Tian T, Yuan S, Shi J, Bedwell GJ, Yang R, Aiken C, Engelman AN, Lusk CP, Lin C, Xiong Y. Shen Q, et al. Nat Struct Mol Biol. 2023 Apr;30(4):425-435. doi: 10.1038/s41594-023-00925-9. Epub 2023 Feb 20. Nat Struct Mol Biol. 2023. PMID: 36807645 Free PMC article.
References
- Fahrenkrog B, Aebi U. The nuclear pore complex: nucleocytoplasmic transport and beyond. Nat Rev Mol Cell Biol. 2003;4:757–766. - PubMed
- Maco B, Fahrenkrog B, Huang NP, Aebi U. Nuclear pore complex structure and plasticity revealed by electron and atomic force microscopy. Methods Mol Biol. 2006;322:273–288. - PubMed
- Alber F, Dokudovskaya S, Veenhoff LM, Zhang W, Kipper J, et al. The molecular architecture of the nuclear pore complex. Nature. 2007;450:695–701. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous