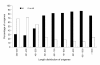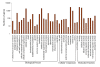De novo assembly and characterization of root transcriptome using Illumina paired-end sequencing and development of cSSR markers in sweet potato (Ipomoea batatas) - PubMed (original) (raw)
De novo assembly and characterization of root transcriptome using Illumina paired-end sequencing and development of cSSR markers in sweet potato (Ipomoea batatas)
Zhangying Wang et al. BMC Genomics. 2010.
Abstract
Background: The tuberous root of sweet potato is an important agricultural and biological organ. There are not sufficient transcriptomic and genomic data in public databases for understanding of the molecular mechanism underlying the tuberous root formation and development. Thus, high throughput transcriptome sequencing is needed to generate enormous transcript sequences from sweet potato root for gene discovery and molecular marker development.
Results: In this study, more than 59 million sequencing reads were generated using Illumina paired-end sequencing technology. De novo assembly yielded 56,516 unigenes with an average length of 581 bp. Based on sequence similarity search with known proteins, a total of 35,051 (62.02%) genes were identified. Out of these annotated unigenes, 5,046 and 11,983 unigenes were assigned to gene ontology and clusters of orthologous group, respectively. Searching against the Kyoto Encyclopedia of Genes and Genomes Pathway database (KEGG) indicated that 17,598 (31.14%) unigenes were mapped to 124 KEGG pathways, and 11,056 were assigned to metabolic pathways, which were well represented by carbohydrate metabolism and biosynthesis of secondary metabolite. In addition, 4,114 cDNA SSRs (cSSRs) were identified as potential molecular markers in our unigenes. One hundred pairs of PCR primers were designed and used for validation of the amplification and assessment of the polymorphism in genomic DNA pools. The result revealed that 92 primer pairs were successfully amplified in initial screening tests.
Conclusion: This study generated a substantial fraction of sweet potato transcript sequences, which can be used to discover novel genes associated with tuberous root formation and development and will also make it possible to construct high density microarrays for further characterization of gene expression profiles during these processes. Thousands of cSSR markers identified in the present study can enrich molecular markers and will facilitate marker-assisted selection in sweet potato breeding. Overall, these sequences and markers will provide valuable resources for the sweet potato community. Additionally, these results also suggested that transcriptome analysis based on Illumina paired-end sequencing is a powerful tool for gene discovery and molecular marker development for non-model species, especially those with large and complex genome.
Figures
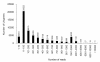
Figure 1
Gap distribution of assembled scaffolds and unigenes. Gap distribution (N/size) %: gap percentage (N amount/sequence length) distribution.
Figure 2
Assessment of assembly quality. Distribution of unique-mapped reads of the assembled unigenes.
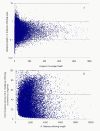
Figure 3
Comparison of I. batatas unigenes to orthologous A. thaliana coding sequences. (A) The ratio of I. batatas unigene length to A. thaliana ortholog length was plotted against I. batatas unigene coverage depth. (B) Total percent of A. thaliana ortholog coding sequence that was covered by all I. batatas unigenes. In total, 502 orthologs could be covered by unigenes with a percentage more than 80%, and the cover percentage of around 5,000 orthologs ranged from 50-80%. Additionally, 27% orthologs were covered with only 20% or lower.
Figure 4
Comparison of unigene length between hit and no-hit unigenes. Longer contigs were more likely to have BLAST matches in protein databases. In this study, 79% of unigenes over 500 bp in length had BLAST matches, whereas only 30% of unigenes shorter than 300 bp did.
Figure 5
Characteristics of similarity search of unigenes against Nr and Swiss-Prot databases. (A) E-value distribution of BLAST hits for each unigene with a cutoff E-value of 1.0E-5 in Nr database. (B) E-value distribution of BLAST hits for each unigene with a cutoff E-value of 1.0E-5 in Swiss-Prot database. (C) Similarity distribution of the top BLAST hits for each unigene in Nr database. (D) Similarity distribution of the top BLAST hits for each unigenes in Swiss-Prot dababase.
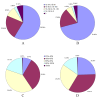
Figure 6
Gene Ontology classification of assembled unigenes. The results are summarized in three main categories: Biological process, Cellular component and Molecular function. In total, 5,046 unigenes with BLAST matches to known proteins were assigned to gene ontology.
Figure 7
Histogram presentation of clusters of orthologous groups (COG) classification. All unigenes were aligned to COG database to predict and classify possible functions. Out of 27,435 Nr hits, 11,983 sequences were assigned to 25 COG classifications.
Figure 8
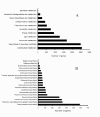
Pathway assignment based on KEGG. (A) Classification based on metabolism categories; (B) Classification based on secondary metabolite categories.
Figure 9
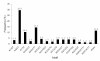
Frequency distribution of cSSRs based on motif sequence types. Within the searched cSSRs, a total of 160 motif sequence types were identified, of which, di-, tri-, tetra-, penta- and hexa-nucleotide repeat existed 4, 10, 30, 57 and 59 types, respectively. The AG/CT di-nucleotide repeat motif was the most abundant motif detected in our cSSRs.
Similar articles
- Transcriptome analysis of the roots at early and late seedling stages using Illumina paired-end sequencing and development of EST-SSR markers in radish.
Wang S, Wang X, He Q, Liu X, Xu W, Li L, Gao J, Wang F. Wang S, et al. Plant Cell Rep. 2012 Aug;31(8):1437-47. doi: 10.1007/s00299-012-1259-3. Epub 2012 Apr 4. Plant Cell Rep. 2012. PMID: 22476438 - Digital gene expression analysis based on integrated de novo transcriptome assembly of sweet potato [Ipomoea batatas (L.) Lam].
Tao X, Gu YH, Wang HY, Zheng W, Li X, Zhao CW, Zhang YZ. Tao X, et al. PLoS One. 2012;7(4):e36234. doi: 10.1371/journal.pone.0036234. Epub 2012 Apr 27. PLoS One. 2012. PMID: 22558397 Free PMC article. - Current status in whole genome sequencing and analysis of Ipomoea spp.
Isobe S, Shirasawa K, Hirakawa H. Isobe S, et al. Plant Cell Rep. 2019 Nov;38(11):1365-1371. doi: 10.1007/s00299-019-02464-4. Epub 2019 Aug 29. Plant Cell Rep. 2019. PMID: 31468128 Free PMC article. Review. - Research Progress in the Mechanisms of Resistance to Biotic Stress in Sweet Potato.
Yang Y, Chen Y, Bo Y, Liu Q, Zhai H. Yang Y, et al. Genes (Basel). 2023 Nov 20;14(11):2106. doi: 10.3390/genes14112106. Genes (Basel). 2023. PMID: 38003049 Free PMC article. Review.
Cited by
- De novo assembly and characterization of transcriptome using Illumina paired-end sequencing and identification of CesA gene in ramie (Boehmeria nivea L. Gaud).
Liu T, Zhu S, Tang Q, Chen P, Yu Y, Tang S. Liu T, et al. BMC Genomics. 2013 Feb 26;14:125. doi: 10.1186/1471-2164-14-125. BMC Genomics. 2013. PMID: 23442184 Free PMC article. - Transcriptome sequencing and analysis of the entomopathogenic fungus Hirsutella sinensis isolated from Ophiocordyceps sinensis.
Liu ZQ, Lin S, Baker PJ, Wu LF, Wang XR, Wu H, Xu F, Wang HY, Brathwaite ME, Zheng YG. Liu ZQ, et al. BMC Genomics. 2015 Feb 21;16(1):106. doi: 10.1186/s12864-015-1269-y. BMC Genomics. 2015. PMID: 25765329 Free PMC article. - De novo transcriptome assembly in chili pepper (Capsicum frutescens) to identify genes involved in the biosynthesis of capsaicinoids.
Liu S, Li W, Wu Y, Chen C, Lei J. Liu S, et al. PLoS One. 2013;8(1):e48156. doi: 10.1371/journal.pone.0048156. Epub 2013 Jan 22. PLoS One. 2013. PMID: 23349661 Free PMC article. - De novo assembly and characterisation of the transcriptome during seed development, and generation of genic-SSR markers in peanut (Arachis hypogaea L.).
Zhang J, Liang S, Duan J, Wang J, Chen S, Cheng Z, Zhang Q, Liang X, Li Y. Zhang J, et al. BMC Genomics. 2012 Mar 12;13:90. doi: 10.1186/1471-2164-13-90. BMC Genomics. 2012. PMID: 22409576 Free PMC article. - De novo transcriptome sequencing and metabolite profiling analyses reveal the complex metabolic genes involved in the terpenoid biosynthesis in Blue Anise Sage (Salvia guaranitica L.).
Ali M, Hussain RM, Rehman NU, She G, Li P, Wan X, Guo L, Zhao J. Ali M, et al. DNA Res. 2018 Dec 1;25(6):597-617. doi: 10.1093/dnares/dsy028. DNA Res. 2018. PMID: 30188980 Free PMC article.
References
- The Food and Agriculture Organization. http://faostat.fao.org/
- Hahn SK. A quantitative approach to source potentials and sink capacities among reciprocal grafts of sweet potato varieties. Crop Sci. 1977;17:559–562. doi: 10.2135/cropsci1977.0011183X001700040020x. - DOI
- Buteler MI, Jarret RL, Labonte DR. Sequence characterization of microsatellites in diploid and polyploid Ipomoea. Theor Appl Genet. 1999;99:123–132. doi: 10.1007/s001220051216. - DOI
- Hu JJ, Nakatani M, Mizuno K, Fujimura T. Development and Characterization of Microsatellite Markers in Sweetpotato. Breeding Science. 2004;54:177–188. doi: 10.1270/jsbbs.54.177. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources