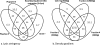Accessing the soil metagenome for studies of microbial diversity - PubMed (original) (raw)
Accessing the soil metagenome for studies of microbial diversity
Tom O Delmont et al. Appl Environ Microbiol. 2011 Feb.
Abstract
Soil microbial communities contain the highest level of prokaryotic diversity of any environment, and metagenomic approaches involving the extraction of DNA from soil can improve our access to these communities. Most analyses of soil biodiversity and function assume that the DNA extracted represents the microbial community in the soil, but subsequent interpretations are limited by the DNA recovered from the soil. Unfortunately, extraction methods do not provide a uniform and unbiased subsample of metagenomic DNA, and as a consequence, accurate species distributions cannot be determined. Moreover, any bias will propagate errors in estimations of overall microbial diversity and may exclude some microbial classes from study and exploitation. To improve metagenomic approaches, investigate DNA extraction biases, and provide tools for assessing the relative abundances of different groups, we explored the biodiversity of the accessible community DNA by fractioning the metagenomic DNA as a function of (i) vertical soil sampling, (ii) density gradients (cell separation), (iii) cell lysis stringency, and (iv) DNA fragment size distribution. Each fraction had a unique genetic diversity, with different predominant and rare species (based on ribosomal intergenic spacer analysis [RISA] fingerprinting and phylochips). All fractions contributed to the number of bacterial groups uncovered in the metagenome, thus increasing the DNA pool for further applications. Indeed, we were able to access a more genetically diverse proportion of the metagenome (a gain of more than 80% compared to the best single extraction method), limit the predominance of a few genomes, and increase the species richness per sequencing effort. This work stresses the difference between extracted DNA pools and the currently inaccessible complete soil metagenome.
Figures
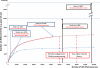
FIG. 1.
Theoretical contribution of the Terragenome Initiative to soil diversity exploration, which starts with 60 “454” titanium plates and the construction of a 2-million-fosmid (40-kb inserts) clone library in the context of soil microbial diversity estimation studies (Metasoil Project). (Based on data from references , , , , and .)
FIG. 2.
Schematic of the different classes of DNA separation methods, starting with physical distance in the field and then density differences in Nycodenz gels, resistance to cell lysis, and finally DNA size separation by pulsed-field gel electrophoresis.
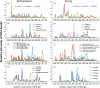
FIG. 3.
Multiple examples of ribosomal intergenic spacer analysis (RISA) electropherograms of DNA from the Ecully and Park Grass, Rothamsted, soils illustrating the differences in the diversities of microbial community DNA as a function of the applied separation technique. Graphs represent relative RISA band intensities as a function of travel time in the gel. (A) Physical separation with MP bead-beating direct DNA extraction of soil samples from different depths (0 to 3, 12 to 15, and 18 to 21 cm deep). (B) Cellular fractionation in a density gradient (Gram-positive lysis after centrifugation at 5,000 × g). (C) Cell lysis with MP bead-beating and MoBio kit direct DNA extractions (depth, 12 to 15 cm) and Epicentre Gram-positive, bead-beating, and DNA tissue indirect DNA extractions (depth, 0 to 10 cm). (D) Metagenomic DNA fractionation by PFGE after extraction by plug protocol B (depth, 10 to 20 cm).
FIG. 4.
Principal component analysis (showing the first and second components) of the matrix data for the RISA analysis from each DNA separation method. The percentages of variance of all axes are shown in the upper left corner. BB, bead beating; A, B, C, D, and E, agarose plug protocols; Bw1, Bw2, and Bw3, low-, medium-, and high-molecular-weight DNA extracted with plug protocol B; M, MoBio Ultraclean kit; G, Epicentre G+ kit; c, cell ring from the Nycodenz density gradient separation; top, DNA recovered from the different fractions above the cell ring. The numbers 1 to 7 refer to the depth intervals (3 cm deep each) of the soil samples from the soil core, with 1 being 0 to 3 cm and 2 being 4 to 6 cm, etc.; 1,000g refers to DNA recovered from the cell ring in the Nycodenz gradient when the centrifuge was operated at 1,000 × g rather than the usual 9,000 × g.
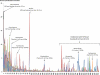
FIG. 5.
Phylogenetic distribution (genus level) of 14 DNA pools for 360 different genera. The genus order is based on the decreasing percentage of those detected in the DNA pool extracted with MP bead-beating direct lysis of the surface (0- to 3-cm) soil sample (black line) from genera 1 to 218. The order from genera 218 to 360 (where the genera were not detected in the reference DNA pool) is alphabetical.
FIG. 6.
Venn diagram showing percentages of probe hybridization coverage (out of over 3,000 total) between DNA extraction protocols as a function of the lysis stringency (a) and location in a Nycodenz density gradient at different centrifugation speeds (b).
FIG. 7.
Rarefaction curve based on phylogenetic microarray analyses of 15 different (based on extraction methods) DNA pools from the Rothamsted soil samples. The percentage of positive probes is plotted against the number of probes tested over multiple microarrays used to test different DNA pools.
Similar articles
- Phylogenetic molecular ecological network of soil microbial communities in response to elevated CO2.
Zhou J, Deng Y, Luo F, He Z, Yang Y. Zhou J, et al. mBio. 2011 Jul 26;2(4):e00122-11. doi: 10.1128/mBio.00122-11. Print 2011. mBio. 2011. PMID: 21791581 Free PMC article. - Soil-specific limitations for access and analysis of soil microbial communities by metagenomics.
Lombard N, Prestat E, van Elsas JD, Simonet P. Lombard N, et al. FEMS Microbiol Ecol. 2011 Oct;78(1):31-49. doi: 10.1111/j.1574-6941.2011.01140.x. Epub 2011 Jun 28. FEMS Microbiol Ecol. 2011. PMID: 21631545 Review. - Microbial biodiversity of meadows under different modes of land use: catabolic and genetic fingerprinting.
Wolinska A, Frąc M, Oszust K, Szafranek-Nakonieczna A, Zielenkiewicz U, Stępniewska Z. Wolinska A, et al. World J Microbiol Biotechnol. 2017 Aug;33(8):154. doi: 10.1007/s11274-017-2318-2. Epub 2017 Jul 5. World J Microbiol Biotechnol. 2017. PMID: 28681284 Free PMC article. - Organic mulching positively regulates the soil microbial communities and ecosystem functions in tea plantation.
Zhang S, Wang Y, Sun L, Qiu C, Ding Y, Gu H, Wang L, Wang Z, Ding Z. Zhang S, et al. BMC Microbiol. 2020 Apr 29;20(1):103. doi: 10.1186/s12866-020-01794-8. BMC Microbiol. 2020. PMID: 32349665 Free PMC article. - From Microscopy to Genomic Approach in Soil Biodiversity Assessment.
Popescu L, Cao ZP. Popescu L, et al. Curr Issues Mol Biol. 2018;27:195-198. doi: 10.21775/cimb.027.195. Epub 2017 Sep 8. Curr Issues Mol Biol. 2018. PMID: 28885183 Review.
Cited by
- Response Characteristics of the Community Structure and Metabolic Genes of Oil-Recovery Bacteria after Targeted Activation of Petroleum Hydrocarbon-Degrading Bacteria in Low-Permeability Oil Reservoirs.
Gao Y, Wang W, Jiang S, Jin Z, Guo M, Wang M, Li H, Cui K. Gao Y, et al. ACS Omega. 2024 Jul 27;9(31):33448-33458. doi: 10.1021/acsomega.3c10334. eCollection 2024 Aug 6. ACS Omega. 2024. PMID: 39130570 Free PMC article. - Non-invasive real-time genomic monitoring of the critically endangered kākāpō.
Urban L, Miller AK, Eason D, Vercoe D, Shaffer M, Wilkinson SP, Jeunen GJ, Gemmell NJ, Digby A. Urban L, et al. Elife. 2023 Dec 28;12:RP84553. doi: 10.7554/eLife.84553. Elife. 2023. PMID: 38153986 Free PMC article. - aKmerBroom: Ancient oral DNA decontamination using Bloom filters on k-mer sets.
Duitama González C, Rangavittal S, Vicedomini R, Chikhi R, Richard H. Duitama González C, et al. iScience. 2023 Sep 29;26(11):108057. doi: 10.1016/j.isci.2023.108057. eCollection 2023 Nov 17. iScience. 2023. PMID: 37876815 Free PMC article. - Siderophore-mediated iron partition promotes dynamical coexistence between cooperators and cheaters.
Shao J, Rong N, Wu Z, Gu S, Liu B, Shen N, Li Z. Shao J, et al. iScience. 2023 Jul 20;26(9):107396. doi: 10.1016/j.isci.2023.107396. eCollection 2023 Sep 15. iScience. 2023. PMID: 37701813 Free PMC article. - Integrating Biochar, Bacteria, and Plants for Sustainable Remediation of Soils Contaminated with Organic Pollutants.
Xiang L, Harindintwali JD, Wang F, Redmile-Gordon M, Chang SX, Fu Y, He C, Muhoza B, Brahushi F, Bolan N, Jiang X, Ok YS, Rinklebe J, Schaeffer A, Zhu YG, Tiedje JM, Xing B. Xiang L, et al. Environ Sci Technol. 2022 Dec 6;56(23):16546-16566. doi: 10.1021/acs.est.2c02976. Epub 2022 Oct 27. Environ Sci Technol. 2022. PMID: 36301703 Free PMC article. Review.
References
- Berry, A. E., C. Chiocchini, T. Selby, M. Sosio, and E. M. Wellington. 2003. Isolation of high molecular weight DNA from soil for cloning into BAC vectors. FEMS Microbiol. Lett. 223:15-20. - PubMed
- Bertrand, H., et al. 2005. High molecular weight DNA recovery from soils prerequisite for biotechnological metagenomic library construction. J. Microbiol. Methods 62:1-11. - PubMed
- Boubakri, H., M. Beuf, P. Simonet, and T. M. Vogel. 2006. Development of metagenomic DNA shuffling for the construction of a xenobiotic gene. Gene 375:87-94. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources