Disordered microbial communities in the upper respiratory tract of cigarette smokers - PubMed (original) (raw)
Disordered microbial communities in the upper respiratory tract of cigarette smokers
Emily S Charlson et al. PLoS One. 2010.
Abstract
Cigarette smokers have an increased risk of infectious diseases involving the respiratory tract. Some effects of smoking on specific respiratory tract bacteria have been described, but the consequences for global airway microbial community composition have not been determined. Here, we used culture-independent high-density sequencing to analyze the microbiota from the right and left nasopharynx and oropharynx of 29 smoking and 33 nonsmoking healthy asymptomatic adults to assess microbial composition and effects of cigarette smoking. Bacterial communities were profiled using 454 pyrosequencing of 16S sequence tags (803,391 total reads), aligned to 16S rRNA databases, and communities compared using the UniFrac distance metric. A Random Forest machine-learning algorithm was used to predict smoking status and identify taxa that best distinguished between smokers and nonsmokers. Community composition was primarily determined by airway site, with individuals exhibiting minimal side-of-body or temporal variation. Within airway habitats, microbiota from smokers were significantly more diverse than nonsmokers and clustered separately. The distributions of several genera were systematically altered by smoking in both the oro- and nasopharynx, and there was an enrichment of anaerobic lineages associated with periodontal disease in the oropharynx. These results indicate that distinct regions of the human upper respiratory tract contain characteristic microbial communities that exhibit disordered patterns in cigarette smokers, both in individual components and global structure, which may contribute to the prevalence of respiratory tract complications in this population.
Conflict of interest statement
Competing Interests: The authors have declared that no competing interests exist.
Figures
Figure 1. Comparison of bacterial community composition reveals that the upper airway microbiota is primarily structured by body habitat.
Unweighted UniFrac was used to generated distances between oropharynx (red), nasopharynx (pink) and fecal (blue) microbiome samples, then scatterplots were generated using Principal Coordinate Analysis. The percentage of variation explained by each PCoA is indicated on the axes. The differences among communities from different body sites was significant with p<0.001 (t-test with permutation). Fecal microbial communities were from .
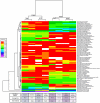
Figure 2. Analysis of abundances of bacterial lineages demonstrates that oro- and nasopharyngeal bacterial communities cluster based on smoking status.
The relative abundance of each genus (rows) is shown by the key to the left of the figure. Communities are clustered by hierarchical clustering using complete linkage of Euclidean distance matrices. The number of times each split in the tree is seen in 1,000 bootstrapped samples is indicated at each node. The tree to the left of the heatmap groups genera together based on similarity of abundance profiles (i.e. if two genera are close in the tree, their abundance profiles across each airway site are similar).
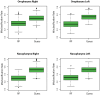
Figure 3. Partitioning airway microbial communities by smoking status using Random Forrest.
Bacterial communities from each airway site were sorted by smoking status using the Random Forests trained algorithm and compared to guessing. Misclassification frequencies are plotted by airway site and side of body. RF = Random Forrest machine. Guess = guessing alone. The lower- and upper-most bars designate the lowest and highest value excluding outliers (defined as >1.5*IQR). The bottom and top of the green boxes denote the lower and upper hinge (close to 25% and 75% quantiles). The heavy black line designates the median misclassification frequency. The distribution of misclassification errors is significantly different between the two algorithms (P – value<2.2E-16 for all airway sites, Friedman Rank Sum test) and in all airway sites, Random Forests performs better than guessing (95% Confidence Interval: oropharynx right (−0.15–−0.13), oropharynx left (−0.20–−0.18); nasopharynx right (−0.23–−0.22), nasopharynx left (−0.22–−0.20).
Similar articles
- Characterization of the upper and lower respiratory tract microbiota in Piedmontese calves.
Nicola I, Cerutti F, Grego E, Bertone I, Gianella P, D'Angelo A, Peletto S, Bellino C. Nicola I, et al. Microbiome. 2017 Nov 21;5(1):152. doi: 10.1186/s40168-017-0372-5. Microbiome. 2017. PMID: 29157308 Free PMC article. - Culture and Molecular Profiling of the Respiratory Tract Microbiota.
Whelan FJ, Rossi L, Stearns JC, Surette MG. Whelan FJ, et al. Methods Mol Biol. 2018;1849:49-61. doi: 10.1007/978-1-4939-8728-3_4. Methods Mol Biol. 2018. PMID: 30298247 - Topographical continuity of bacterial populations in the healthy human respiratory tract.
Charlson ES, Bittinger K, Haas AR, Fitzgerald AS, Frank I, Yadav A, Bushman FD, Collman RG. Charlson ES, et al. Am J Respir Crit Care Med. 2011 Oct 15;184(8):957-63. doi: 10.1164/rccm.201104-0655OC. Epub 2011 Jun 16. Am J Respir Crit Care Med. 2011. PMID: 21680950 Free PMC article. - The loss of topography in the microbial communities of the upper respiratory tract in the elderly.
Whelan FJ, Verschoor CP, Stearns JC, Rossi L, Luinstra K, Loeb M, Smieja M, Johnstone J, Surette MG, Bowdish DM. Whelan FJ, et al. Ann Am Thorac Soc. 2014 May;11(4):513-21. doi: 10.1513/AnnalsATS.201310-351OC. Ann Am Thorac Soc. 2014. PMID: 24601676 - The Microbiome and the Respiratory Tract.
Dickson RP, Erb-Downward JR, Martinez FJ, Huffnagle GB. Dickson RP, et al. Annu Rev Physiol. 2016;78:481-504. doi: 10.1146/annurev-physiol-021115-105238. Epub 2015 Nov 2. Annu Rev Physiol. 2016. PMID: 26527186 Free PMC article. Review.
Cited by
- A metagenomic approach to characterization of the vaginal microbiome signature in pregnancy.
Aagaard K, Riehle K, Ma J, Segata N, Mistretta TA, Coarfa C, Raza S, Rosenbaum S, Van den Veyver I, Milosavljevic A, Gevers D, Huttenhower C, Petrosino J, Versalovic J. Aagaard K, et al. PLoS One. 2012;7(6):e36466. doi: 10.1371/journal.pone.0036466. Epub 2012 Jun 13. PLoS One. 2012. PMID: 22719832 Free PMC article. - Higher rates of Clostridium difficile infection among smokers.
Rogers MA, Greene MT, Saint S, Chenoweth CE, Malani PN, Trivedi I, Aronoff DM. Rogers MA, et al. PLoS One. 2012;7(7):e42091. doi: 10.1371/journal.pone.0042091. Epub 2012 Jul 27. PLoS One. 2012. PMID: 22848714 Free PMC article. - The lung mycobiome: an emerging field of the human respiratory microbiome.
Nguyen LD, Viscogliosi E, Delhaes L. Nguyen LD, et al. Front Microbiol. 2015 Feb 13;6:89. doi: 10.3389/fmicb.2015.00089. eCollection 2015. Front Microbiol. 2015. PMID: 25762987 Free PMC article. - The role of the local microbial ecosystem in respiratory health and disease.
de Steenhuijsen Piters WA, Sanders EA, Bogaert D. de Steenhuijsen Piters WA, et al. Philos Trans R Soc Lond B Biol Sci. 2015 Aug 19;370(1675):20140294. doi: 10.1098/rstb.2014.0294. Philos Trans R Soc Lond B Biol Sci. 2015. PMID: 26150660 Free PMC article. Review. - Upper Respiratory Dysbiosis with a Facultative-dominated Ecotype in Advanced Lung Disease and Dynamic Change after Lung Transplant.
Simon-Soro A, Sohn MB, McGinniss JE, Imai I, Brown MC, Knecht VR, Bailey A, Clarke EL, Cantu E, Li H, Bittinger K, Diamond JM, Christie JD, Bushman FD, Collman RG. Simon-Soro A, et al. Ann Am Thorac Soc. 2019 Nov;16(11):1383-1391. doi: 10.1513/AnnalsATS.201904-299OC. Ann Am Thorac Soc. 2019. PMID: 31415219 Free PMC article.
References
- Aronson MD, Weiss ST, Ben RL, Komaroff AL. Association between cigarette smoking and acute respiratory tract illness in young adults. JAMA. 1982;248:181–183. - PubMed
- Turkeltaub PC, Gergen PJ. Prevalence of upper and lower respiratory conditions in the US population by social and environmental factors: data from the second National Health and Nutrition Examination Survey, 1976 to 1980 (NHANES II). Ann Allergy. 1991;67:147–154. - PubMed
- Brook I, Gober AE. Recovery of potential pathogens in the nasopharynx of healthy and otitis media-prone children and their smoking and nonsmoking parents. Ann Otol Rhinol Laryngol. 2008;117:727–730. - PubMed
- El Ahmer OR, Essery SD, Saadi AT, Raza MW, Ogilvie MM, et al. The effect of cigarette smoke on adherence of respiratory pathogens to buccal epithelial cells. FEMS Immunol Med Microbiol. 1999;23:27–36. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases