Identification and characterization of nuclear pore complex components in Arabidopsis thaliana - PubMed (original) (raw)
Identification and characterization of nuclear pore complex components in Arabidopsis thaliana
Kentaro Tamura et al. Plant Cell. 2010 Dec.
Abstract
The nuclear pore complex (NPC) facilitates nucleocytoplasmic transport, a crucial process for various cellular activities. The NPC comprises ~30 nucleoporins and is well characterized in vertebrates and yeast. However, only eight plant nucleoporins have been identified, and little information is available about the complete molecular structure of plant NPCs. In this study, an interactive proteomic approach was used to identify Arabidopsis thaliana nucleoporins. A series of five cycles of interactive proteomic analysis was performed using green fluorescent protein (GFP)-tagged nucleoporins. The identified nucleoporins were then cloned and subcellular localization analyses were performed. We found that the plant NPC contains at least 30 nucleoporins, 22 of which had not been previously annotated. Surprisingly, plant nucleoporins shared a similar domain organization to their vertebrate (human) and yeast (Saccharomyces cerevisiae) counterparts. Moreover, the plant nucleoporins exhibited higher sequence homology to vertebrate nucleoporins than to yeast nucleoporins. Plant NPCs lacked seven components (NUCLEOPORIN358 [Nup358], Nup188, Nup153, Nup45, Nup37, NUCLEAR DIVISION CYCLE1, and PORE MEMBRANE PROTEIN OF 121 kD) that were present in vertebrate NPCs. However, plants possessed a nucleoporin, Nup136/Nup1, that contained Phe-Gly repeats, and sequence analysis failed to identify a vertebrate homolog for this protein. Interestingly, Nup136-GFP showed greater mobility on the nuclear envelope than did other nucleoporins, and a Nup136/Nup1 deficiency caused various defects in plant development. These findings provide valuable new information about plant NPC structure and function.
Figures
Figure 1.
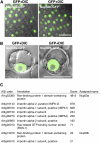
Arabidopsis Nucleoporins That Coimmunoprecipitate with RAE1-GFP in Transgenic Arabidopsis Plants. (A) Root tip cells from transgenic Arabidopsis plants stably expressing RAE1-GFP. The images were obtained by confocal laser scanning microscopy. Bar = 20 μm. (B) The nuclear surface of a root cell from transgenic Arabidopsis plants stably expressing RAE1-GFP. The image was obtained by confocal laser scanning microscopy. Bar = 10 μm. (C) and (D) Silver staining (C) and immunoblot analysis (D) of lysates from wild-type Arabidopsis (wt) and transgenic (RAE1-GFP) plants immunoprecipitated with anti-GFP antibody. Immunoprecipitates were separated by SDS-PAGE followed by silver staining (C) or immunoblot analysis with an anti-GFP antibody (D). Asterisks indicate nonspecific signals. (E) Identification of Arabidopsis nucleoporins by mass spectrometry. Extracts from transgenic Arabidopsis plants expressing RAE1-GFP were immunoprecipitated with anti-GFP antibody and then subjected to LTQ-Orbitrap mass spectrometry. Arabidopsis Genome Initiative (AGI) codes and annotations were obtained from the TAIR database (
). Scores were calculated by Mascot (Matrix Science). We assigned a name to each nucleoporin candidate identified in this study.
Figure 2.
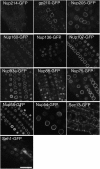
Subcellular Localization of Stably Expressed GFP-Tagged Nucleoporins in Arabidopsis. GFP fusion constructs were generated for 13 nucleoporins identified after anti-GFP antibody immunoprecipitation of a detergent-solubilized fraction from RAE1-GFP plants (Figure 1D). The constructs were then stably expressed in Arabidopsis plants. Images of epidermal root tip cells were all obtained at the same magnification using confocal laser scanning microscopy. Bar = 20 μm.
Figure 3.
Identification of Arabidopsis Nup50a and Nup50b. (A) and (B) Fluorescence images of root tip cells from transgenic plants stably expressing Nup50a-GFP (left panel) or Nup50b-GFP (right panel) (A). Fluorescence images of protoplasts transiently expressing Nup50a-GFP (left panel) or Nup50b-GFP (right panel) (B). Images were obtained by confocal laser scanning microscopy (GFP) and differential interference contrast (DIC) microscopy. Fluorescence–differential interference contrast overlays are shown. Bars = 20 μm. (C) Mass spectrometry identification of Arabidopsis nucleoporins and nucleocytoplasmic transport factors that interact with Nup50a-GFP. Immunoprecipitates from transgenic Arabidopsis plants expressing Nup50a-GFP were subjected to LTQ-Orbitrap mass spectrometry. Arabidopsis Genome Initiative (AGI) codes and annotations were obtained from the TAIR database (
). Scores were calculated by Mascot. Assigned names represent nucleoporins identified and named in this study.
Figure 4.
Schematic Representation of the Predicted Domain Features of Arabidopsis Nucleoporins. The 30 Arabidopsis nucleoporins separate into four groups: (1) FG-bearing nucleoporins (FG-Nups); (2) Nup107-160 subcomplex proteins, corresponding to the outer ring structure of the NPC; (3) Nup93 subcomplex proteins, corresponding to the inner ring structure of the NPC; and (4) proteins with other functions. The scale at the top indicates the number of amino acids. A single orange vertical line in FG-Nups indicates the position of an FG repeat. The Max scores and E-values indicate sequence homologies with human (Homo sapiens) and yeast (Saccharomyces cerevisiae) counterparts. Different names for S. cerevisiae homologs are shown in parentheses. No homologs of Nup43, ALADIN, gp210, and Elys/HOS1 were identified in S. cerevisiae. Procedures outlining domain prediction are described in Methods, and details of the domains are shown in
Supplemental Table 2
online. AGI, Arabidopsis Genome Initiative.
Figure 5.
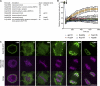
Nup136/Nup1 Is a Nucleoporin with Dynamic Behavior on the Nuclear Envelope. (A) Mass spectrometry identification of Arabidopsis nucleoporins and nucleocytoplasmic transport factors that interact with Nup136-GFP. Immunoprecipitates from transgenic Arabidopsis plants expressing Nup136-GFP were subjected to LTQ-Orbitrap mass spectrometry. Arabidopsis Genome Initiative (AGI) codes and annotations were obtained from the TAIR database (
). Scores were calculated by Mascot. Assigned names represent nucleoporins identified and named in this study. (B) FRAP analysis of nucleoporin behavior on the nuclear envelope. Constructs encoding GFP fusions with gp210, Nup93a, Nup136, Nup54, Nup107, and Nup88 were stably expressed in tobacco BY-2 cells. An arrow indicates the photobleaching time point. Vertical bars represent
sd
(n ≥ 6). (C) Cultured tobacco cells were cotransformed with constructs encoding Nup136-GFP and Histone2B-tdTomato to visualize the nucleoporin (green, top panels) and chromatin DNA (magenta, bottom panels), respectively. Living tobacco cultured cells were observed during cell division. The confocal time-lapse images (single planes) are shown from left to right, and the time is shown in seconds after breakdown of the nuclear envelope. Bar = 10 μm.
Figure 6.
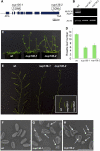
Two Knockout Mutants, nup136-1 and nup136-2, Exhibit Various Defects in Plant Development. (A) A schematic representation of the Nup136/Nup1 gene, which contains nine exons. The positions of T-DNA insertions in nup136-1 and nup136-2 are shown. Closed boxes represent exons and solid lines represent introns. (B) RT-PCR analysis of NUP136/NUP1 and Actin2 (ACT2) transcripts in wild-type (WT), nup136-1, and nup136-2 cells. Full-length NUP136 transcripts were amplified by PCR. Amplification of NUP136/NUP1 and ACT2 required 30 and 27 PCR cycles, respectively. RT-PCR products were visualized with ethidium bromide. These data are representative of two biological replicates and two technical replicates. (C) Four-week-old wild-type (wt), nup136-1, and _nup136-_2 plants. (D) Flowering time was evaluated by the number of primary rosette leaves formed prior to flowering under long-day conditions. Data represent means ±
sd
(n ≥ 14). (E) The two nup136/nup1 knockout mutants exhibited a defect in fruit maturation. Inset provides a magnified view of representative siliques of the same age. (F) to (H) Scanning electron micrographs of pollen grains of wild-type (F), nup136-1 (G), and nup136-2 (H) plants. Arrows indicate the shrunken pollen grains of the mutants. Bar = 50 μm.
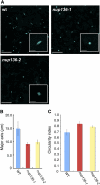
Figure 7.
Effects of a NUP136/NUP1 Knockout on Nuclear Morphology. (A) Nuclear morphology of rosette leaf epidermal cells stained with Hoechst 33342. Images were obtained by confocal laser scanning microscopy. Insets show magnified images of nuclei. Bars = 20 μm. Bars in insets = 10 μm. wt, wild type. (B) and (C) The major axis length (B) and circularity index (C) of nuclei were measured in wild-type, nup136-1, and nup136-2 cells. Data represent means ±
sd
(n ≥ 63).
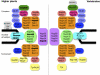
Figure 8.
Comparison between the NPCs of Higher Plants and Vertebrates. Subcomplexes are shown as single units. Plant nucleoporins that were not identified in this study are crossed out. WIT, WIP, and RanGAP were identified previously (Xu et al., 2007a; Zhao et al., 2008).
Similar articles
- Involvement of the nuclear pore complex in morphology of the plant nucleus.
Tamura K, Hara-Nishimura I. Tamura K, et al. Nucleus. 2011 May-Jun;2(3):168-72. doi: 10.4161/nucl.2.3.16175. Nucleus. 2011. PMID: 21818409 Free PMC article. - Nup82 functions redundantly with Nup136 in a salicylic acid-dependent defense response of Arabidopsis thaliana.
Tamura K, Fukao Y, Hatsugai N, Katagiri F, Hara-Nishimura I. Tamura K, et al. Nucleus. 2017 May 4;8(3):301-311. doi: 10.1080/19491034.2017.1279774. Epub 2017 Jan 10. Nucleus. 2017. PMID: 28071978 Free PMC article. - Arabidopsis homolog of the yeast TREX-2 mRNA export complex: components and anchoring nucleoporin.
Lu Q, Tang X, Tian G, Wang F, Liu K, Nguyen V, Kohalmi SE, Keller WA, Tsang EW, Harada JJ, Rothstein SJ, Cui Y. Lu Q, et al. Plant J. 2010 Jan;61(2):259-70. doi: 10.1111/j.1365-313X.2009.04048.x. Epub 2009 Oct 16. Plant J. 2010. PMID: 19843313 - The molecular architecture of the plant nuclear pore complex.
Tamura K, Hara-Nishimura I. Tamura K, et al. J Exp Bot. 2013 Feb;64(4):823-32. doi: 10.1093/jxb/ers258. Epub 2012 Sep 17. J Exp Bot. 2013. PMID: 22987840 Review. - Structure and Assembly of the Nuclear Pore Complex.
Hampoelz B, Andres-Pons A, Kastritis P, Beck M. Hampoelz B, et al. Annu Rev Biophys. 2019 May 6;48:515-536. doi: 10.1146/annurev-biophys-052118-115308. Epub 2019 Apr 3. Annu Rev Biophys. 2019. PMID: 30943044 Review.
Cited by
- Plant nuclear shape is independently determined by the SUN-WIP-WIT2-myosin XI-i complex and CRWN1.
Zhou X, Groves NR, Meier I. Zhou X, et al. Nucleus. 2015;6(2):144-53. doi: 10.1080/19491034.2014.1003512. Epub 2015 Mar 11. Nucleus. 2015. PMID: 25759303 Free PMC article. - Structural and functional studies of the 252 kDa nucleoporin ELYS reveal distinct roles for its three tethered domains.
Bilokapic S, Schwartz TU. Bilokapic S, et al. Structure. 2013 Apr 2;21(4):572-80. doi: 10.1016/j.str.2013.02.006. Epub 2013 Mar 14. Structure. 2013. PMID: 23499022 Free PMC article. - A simple thermodynamic description of phase separation of Nup98 FG domains.
Ng SC, Görlich D. Ng SC, et al. Nat Commun. 2022 Oct 18;13(1):6172. doi: 10.1038/s41467-022-33697-9. Nat Commun. 2022. PMID: 36257947 Free PMC article. - Importin α: functions as a nuclear transport factor and beyond.
Oka M, Yoneda Y. Oka M, et al. Proc Jpn Acad Ser B Phys Biol Sci. 2018;94(7):259-274. doi: 10.2183/pjab.94.018. Proc Jpn Acad Ser B Phys Biol Sci. 2018. PMID: 30078827 Free PMC article. Review. - Modeling cell biological features of meiotic chromosome pairing to study interlock resolution.
Navarro EJ, Marshall WF, Fung JC. Navarro EJ, et al. PLoS Comput Biol. 2022 Jun 13;18(6):e1010252. doi: 10.1371/journal.pcbi.1010252. eCollection 2022 Jun. PLoS Comput Biol. 2022. PMID: 35696428 Free PMC article.
References
- Allen J.L., Douglas M.G. (1989). Organization of the nuclear pore complex in Saccharomyces cerevisiae. J. Ultrastruct. Mol. Struct. Res. 102: 95–108 - PubMed
- Blower M.D., Nachury M., Heald R., Weis K. (2005). A Rae1-containing ribonucleoprotein complex is required for mitotic spindle assembly. Cell 121: 223–234 - PubMed
- Boavida L.C., McCormick S. (2007). Temperature as a determinant factor for increased and reproducible in vitro pollen germination in Arabidopsis thaliana. Plant J. 52: 570–582 - PubMed
- Bodoor K., Shaikh S., Salina D., Raharjo W.H., Bastos R., Lohka M., Burke B. (1999). Sequential recruitment of NPC proteins to the nuclear periphery at the end of mitosis. J. Cell Sci. 112: 2253–2264 - PubMed
- Boisnard-Lorig C., Colon-Carmona A., Bauch M., Hodge S., Doerner P., Bancharel E., Dumas C., Haseloff J., Berger F. (2001). Dynamic analyses of the expression of the HISTONE:YFP fusion protein in Arabidopsis show that syncytial endosperm is divided in mitotic domains. Plant Cell 13: 495–509 - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous