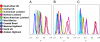Genetic signals of origin, spread, and introgression in a large sample of maize landraces - PubMed (original) (raw)
Genetic signals of origin, spread, and introgression in a large sample of maize landraces
Joost van Heerwaarden et al. Proc Natl Acad Sci U S A. 2011.
Abstract
The last two decades have seen important advances in our knowledge of maize domestication, thanks in part to the contributions of genetic data. Genetic studies have provided firm evidence that maize was domesticated from Balsas teosinte (Zea mays subspecies parviglumis), a wild relative that is endemic to the mid- to lowland regions of southwestern Mexico. An interesting paradox remains, however: Maize cultivars that are most closely related to Balsas teosinte are found mainly in the Mexican highlands where subspecies parviglumis does not grow. Genetic data thus point to primary diffusion of domesticated maize from the highlands rather than from the region of initial domestication. Recent archeological evidence for early lowland cultivation has been consistent with the genetics of domestication, leaving the issue of the ancestral position of highland maize unresolved. We used a new SNP dataset scored in a large number of accessions of both teosinte and maize to take a second look at the geography of the earliest cultivated maize. We found that gene flow between maize and its wild relatives meaningfully impacts our inference of geographic origins. By analyzing differentiation from inferred ancestral gene frequencies, we obtained results that are fully consistent with current ecological, archeological, and genetic data concerning the geography of early maize cultivation.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
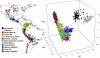
Fig. 1.
(A) Map of sampled maize accessions colored by genetic group. (B) First three genetic PCs of all sampled accessions.
Fig. 2.
(Lower) Bar plot of assignment values for the sample of Mexican accessions: Mexicana (red), parviglumis (green), and mays (blue). (Upper) The solid black line indicates the altitude for each sample. The dotted line marks the minimum altitude at which mexicana occurs.
Fig. 3.
Posterior densities of the genetic drift parameter F for 10 genetic groups with respect to (A) mexicana and (B) parviglumis. Only lowland accessions of the West Mexico group (light blue) were included. (C) Drift of all 10 genetic groups with respect to inferred ancestral frequencies. Light blue represents West Mexico; dotted line indicates the division between lowlands (<1,500 m, solid line) and highlands (>2,000 m).
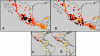
Fig. 4.
Heat maps showing the amount of drift, F, away from (A) observed parviglumis allelic frequencies and (B) mean estimated ancestral frequencies. Each point is based on spatial estimation of current allele frequencies. The colors of the dots range from red for low values to white for high values of F. Black dots mark the lower 0.05 quantile. Upper panels A and B show enlarged sections of the lower panels A and B, respectively.
Similar articles
- Domestication and lowland adaptation of coastal preceramic maize from Paredones, Peru.
Vallebueno-Estrada M, Hernández-Robles GG, González-Orozco E, Lopez-Valdivia I, Rosales Tham T, Vásquez Sánchez V, Swarts K, Dillehay TD, Vielle-Calzada JP, Montiel R. Vallebueno-Estrada M, et al. Elife. 2023 Apr 18;12:e83149. doi: 10.7554/eLife.83149. Elife. 2023. PMID: 37070964 Free PMC article. - The relevance of gene flow with wild relatives in understanding the domestication process.
Moreno-Letelier A, Aguirre-Liguori JA, Piñero D, Vázquez-Lobo A, Eguiarte LE. Moreno-Letelier A, et al. R Soc Open Sci. 2020 Apr 15;7(4):191545. doi: 10.1098/rsos.191545. eCollection 2020 Apr. R Soc Open Sci. 2020. PMID: 32431864 Free PMC article. - Population genomics of Zea species identifies selection signatures during maize domestication and adaptation.
Xu G, Zhang X, Chen W, Zhang R, Li Z, Wen W, Warburton ML, Li J, Li H, Yang X. Xu G, et al. BMC Plant Biol. 2022 Feb 18;22(1):72. doi: 10.1186/s12870-022-03427-w. BMC Plant Biol. 2022. PMID: 35180846 Free PMC article. - Genomic screening for artificial selection during domestication and improvement in maize.
Yamasaki M, Wright SI, McMullen MD. Yamasaki M, et al. Ann Bot. 2007 Nov;100(5):967-73. doi: 10.1093/aob/mcm173. Epub 2007 Aug 18. Ann Bot. 2007. PMID: 17704539 Free PMC article. Review. - Maize domestication and gene interaction.
Stitzer MC, Ross-Ibarra J. Stitzer MC, et al. New Phytol. 2018 Oct;220(2):395-408. doi: 10.1111/nph.15350. Epub 2018 Jul 23. New Phytol. 2018. PMID: 30035321 Review.
Cited by
- Joint analysis of days to flowering reveals independent temperate adaptations in maize.
Swarts K, Bauer E, Glaubitz JC, Ho T, Johnson L, Li Y, Li Y, Miller Z, Romay C, Schön CC, Wang T, Zhang Z, Buckler ES, Bradbury P. Swarts K, et al. Heredity (Edinb). 2021 Jun;126(6):929-941. doi: 10.1038/s41437-021-00422-z. Epub 2021 Apr 22. Heredity (Edinb). 2021. PMID: 33888874 Free PMC article. - A footprint of past climate change on the diversity and population structure of Miscanthus sinensis.
Clark LV, Brummer JE, Głowacka K, Hall MC, Heo K, Peng J, Yamada T, Yoo JH, Yu CY, Zhao H, Long SP, Sacks EJ. Clark LV, et al. Ann Bot. 2014 Jul;114(1):97-107. doi: 10.1093/aob/mcu084. Epub 2014 Jun 10. Ann Bot. 2014. PMID: 24918203 Free PMC article. - Genetic diversity and population structure of native maize populations in Latin America and the Caribbean.
Bedoya CA, Dreisigacker S, Hearne S, Franco J, Mir C, Prasanna BM, Taba S, Charcosset A, Warburton ML. Bedoya CA, et al. PLoS One. 2017 Apr 12;12(4):e0173488. doi: 10.1371/journal.pone.0173488. eCollection 2017. PLoS One. 2017. PMID: 28403177 Free PMC article. - De Novo Genome Assembly of the Japanese Wheat Cultivar Norin 61 Highlights Functional Variation in Flowering Time and Fusarium-Resistant Genes in East Asian Genotypes.
Shimizu KK, Copetti D, Okada M, Wicker T, Tameshige T, Hatakeyama M, Shimizu-Inatsugi R, Aquino C, Nishimura K, Kobayashi F, Murata K, Kuo T, Delorean E, Poland J, Haberer G, Spannagl M, Mayer KFX, Gutierrez-Gonzalez J, Muehlbauer GJ, Monat C, Himmelbach A, Padmarasu S, Mascher M, Walkowiak S, Nakazaki T, Ban T, Kawaura K, Tsuji H, Pozniak C, Stein N, Sese J, Nasuda S, Handa H. Shimizu KK, et al. Plant Cell Physiol. 2021 Mar 25;62(1):8-27. doi: 10.1093/pcp/pcaa152. Plant Cell Physiol. 2021. PMID: 33244607 Free PMC article. - A systems approach to a spatio-temporal understanding of the drought stress response in maize.
Miao Z, Han Z, Zhang T, Chen S, Ma C. Miao Z, et al. Sci Rep. 2017 Jul 26;7(1):6590. doi: 10.1038/s41598-017-06929-y. Sci Rep. 2017. PMID: 28747711 Free PMC article.
References
- Beadle GW. Teosinte and the origin of maize. J Hered. 1939;30:245–247.
- Mangelsdorf PC, Macneish RS, Galinat WC. Domestication of corn. Science. 1964;143:538–545. - PubMed
- Burger JC, Chapman MA, Burke JM. Molecular insights into the evolution of crop plants. Am J Bot. 2008;95:113–122. - PubMed
- Doebley JF, Goodman MM, Stuber CW. Isoenzymatic variation in Zea (Graminae) Syst Bot. 1984;9:203–218.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources