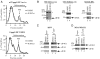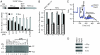Reversing chemoresistance by small molecule inhibition of the translation initiation complex eIF4F - PubMed (original) (raw)
. 2011 Jan 18;108(3):1046-51.
doi: 10.1073/pnas.1011477108. Epub 2010 Dec 29.
David R Hall, Francis Robert, Yuhong Du, Jaeki Min, Lian Li, Min Qui, Iestyn Lewis, Serdar Kurtkaya, Ray Dingledine, Haian Fu, Dima Kozakov, Sandor Vajda, Jerry Pelletier
Affiliations
- PMID: 21191102
- PMCID: PMC3024666
- DOI: 10.1073/pnas.1011477108
Reversing chemoresistance by small molecule inhibition of the translation initiation complex eIF4F
Regina Cencic et al. Proc Natl Acad Sci U S A. 2011.
Erratum in
- Proc Natl Acad Sci U S A. 2011 Apr 19;108(16):6689
Abstract
Deregulation of cap-dependent translation is associated with cancer initiation and progression. The rate-limiting step of protein synthesis is the loading of ribosomes onto mRNA templates stimulated by the heterotrimeric complex, eukaryotic initiation factor (eIF)4F. This step represents an attractive target for anticancer drug discovery because it resides at the nexus of the TOR signaling pathway. We have undertaken an ultra-high-throughput screen to identify inhibitors that prevent assembly of the eIF4F complex. One of the identified compounds blocks interaction between two subunits of eIF4F. As a consequence, cap-dependent translation is inhibited. This compound can reverse tumor chemoresistance in a genetically engineered lymphoma mouse model by sensitizing cells to the proapoptotic action of DNA damage. Molecular modeling experiments provide insight into the mechanism of action of this small molecule inhibitor. Our experiments validate targeting the eIF4F complex as a strategy for cancer therapy to modulate chemosensitivity.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Fig. 1.
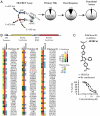
UltraHTS TR-FRET identifies inhibitors of eIF4E:eIF4GI interaction. A. Schematic diagram of TR-FRET based assay and attrition rate from secondary and functional assays. B. Schematic representation of the FF/HCV/Ren bicistronic construct used to characterize inhibitory potential of primary hits in in vitro translation extracts. Heat map showing results of secondary in vitro translation assay and of the counterscreen to identify compounds that nonspecifically inhibited luc enzyme activity. Experiments were performed in duplicates and average values obtained were normalized to DMSO controls. nd, not done. C. Structure of 4E1RCat and its dose response in the TR-FRET assay.
Fig. 2.
Modeling of 4E1RCat bound to eIF4E. A. Location of the largest hot spots of eIF4E. Results are shown for the eIF4E structure cocrystallized with segment 47-66 of 4E-BP1 (PDB code 1WKW), but are essentially identical for the other two available X-ray structures (PDB codes 1IPB and 2W97). The largest consensus site, CS1, (shown in yellow), binds 24 probe clusters and defines the main hot spot. The other large consensus sites are CS2 (magenta, 22 probe clusters), CS3 (cyan, 19 probe clusters), and CS4 (salmon, 10 probe clusters). Consensus site CS6 is small (ochre, 5 probe clusters), but indicates a shallow channel connecting the consensus sites CS1 and CS3. The close-up of the hot spots also shows (in red) the location of residues V69, L131, and I138 on the surface of eIF4E. B. The most likely binding pose of 4E1RCat. The predicted hot spots are superimposed for reference. C. Segment 47-66 of 4E-BP1 from the eIF4E:4E-BP1 complex (PDB code 1WKW) superimposed on the hot spots. For 4E-BP1, the side chain of Y54 of the motif extends toward CS2, L59 is deep in the pocket that binds CS1, confirming the importance of the main hot spot, and the side chain of M60 overlaps with CS6.
Fig. 3.
4E1RCat inhibits cap-dependent translation initiation. A. Inhibition of cap-dependent 80S complex formation by 4E1RCat. 32P-labeled m7GpppG-FF Luc A+ or GpppG-HCV mRNA was incubated with cycloheximide (CHX) and either vehicle (1% DMSO) or 50 μM 4E1RCat in RRL. Total counts recovered from each gradient and the percent mRNA bound in 80S complexes were—m7GpppG-FF/mRNA + 1% DMSO [58895 cpm, 14.2% binding], m7GpppG-FF/mRNA + 4E1RCat [60503 cpm, 10.6% binding], GpppG-HCV/mRNA + 1% DMSO [64426 cpm, 14.2% binding], and GpppG-HCV/mRNA + 4E1RCat [67592 cpm, 17% binding]. B. Effect of 4E1RCat on the interaction between eIF4E and GST-eIF4GI517–606 (left), GST-eIF4GII555–658 (center), and GST-4E-BP1 (right). GST-pull downs were performed in the absence (lanes 1, 3, and 5) or presence (lanes 2, 4, and 6) of 4E1RCat. Glutathionine eluents were probed for the presence of GST-tagged proteins (denoted by an asterisk) and eIF4E by Western blotting. C. 4E1RCat inhibits eIF4F complex assembly in vitro. Pull-down experiments from RSW were performed as described in the Materials and Methods. GTP and m7GTP eluents were fractionated by SDS-PAGE and probed for the presence of eIF4E, eIF4A, and eIF4GI by Western blotting. D. 4E1RCat inhibits eIF4F complex assembly in vivo. Pull-down experiments from cell extracts were performed as described in the Materials and Methods. Input and m7GTP eluents were analyzed by SDS-PAGE and probed for the presence of eIF4E and eIF4GI by Western blotting.
Fig. 4.
Effects of 4E1RCat on translation. A. Effect of 4E1RCat on in vitro translations performed in Krebs extracts programmed with FF/HCV/Ren. A schematic representation of FF/HCV/Ren mRNA is provided (top). In vitro translations were performed in the presence of 35S-Met and a representative autoradiograph of the products after fractionation by 10% SDS-PAGE is provided (bottom). Reactions contained vehicle (1% DMSO) (lane 1), 500 μM m7GDP (lane 2), 500 μM GDP (lane 3), and 50 μM anisomycin (lane 4), the indicated concentrations of 4E1RCat (lanes 5–10), or lacked input mRNA (lane 11). Center: FF and Ren RLU values (relative to DMSO controls) from two independent experiments with the SEM indicated. B. 4E1RCat inhibits protein synthesis in vivo. The rate of incorporation of each radioisotope tracer into TCA-insoluble material was monitored and is expressed relative to vehicle (DMSO) treated cells, which is set at 1. Results are the average of three experiments with the error of the mean shown. C. Polysome profiling analysis of Jurkat cells treated with 50 μM 4E1RCat. D. 4E1RCat inhibits c-Myc and Mcl-1 production. Jurkat cells were treated with 4E1RCat for 1 h, cell extracts prepared, and analyzed by Western blotting for c-Myc (Santa Cruz SC-40), Mcl-1 (Rockland), and actin (Sigma A5441) expression.
Fig. 5.
4E1RCat alters chemosensitivity of Pten+/-Eμ-Myc tumors in vivo. A. Representation of Eμ-Myc model and treatment response. B. Kaplan-Meier plot showing tumor-free survival of mice bearing Pten+/-E_μ_-Myc tumors following treatment with doxorubicin (Dxr, solid black line; n = 10), rapamycin (Rap, solid green line; n = 9), Rap and Dxr (dashed black line; n = 10), 4E1RCat (4E1RCat, solid red line; n = 10), or 4E1RCat and Dxr (dashed red line; n = 10). C. Combination treatment of 4E1RCat and doxorubicin increases the percentage of apoptotic cells. Representative TUNEL staining on sections of Pten+/-E_μ_-Myc tumors following treatments (original magnification × 20-fold). The percentage of cells that stained positive represents the average of four different fields, where 500 cells were counted per field. D. 4E1RCat inhibits Mcl-1 production in vivo. E. 4E1RCat inhibits protein synthesis in vivo.
Similar articles
- Translation initiation complex eIF4F is a therapeutic target for dual mTOR kinase inhibitors in non-Hodgkin lymphoma.
Demosthenous C, Han JJ, Stenson MJ, Maurer MJ, Wellik LE, Link B, Hege K, Dogan A, Sotomayor E, Witzig T, Gupta M. Demosthenous C, et al. Oncotarget. 2015 Apr 20;6(11):9488-501. doi: 10.18632/oncotarget.3378. Oncotarget. 2015. PMID: 25839159 Free PMC article. - Homogenous time resolved fluorescence assay to identify modulators of cap-dependent translation initiation.
Cencic R, Yan Y, Pelletier J. Cencic R, et al. Comb Chem High Throughput Screen. 2007 Mar;10(3):181-8. doi: 10.2174/138620707780126688. Comb Chem High Throughput Screen. 2007. PMID: 17346117 - RNA-tethering assay and eIF4G:eIF4A obligate dimer design uncovers multiple eIF4F functional complexes.
Robert F, Cencic R, Cai R, Schmeing TM, Pelletier J. Robert F, et al. Nucleic Acids Res. 2020 Sep 4;48(15):8562-8575. doi: 10.1093/nar/gkaa646. Nucleic Acids Res. 2020. PMID: 32749456 Free PMC article. - eIF4F: a retrospective.
Merrick WC. Merrick WC. J Biol Chem. 2015 Oct 2;290(40):24091-9. doi: 10.1074/jbc.R115.675280. Epub 2015 Aug 31. J Biol Chem. 2015. PMID: 26324716 Free PMC article. Review. - Attacking a nexus of the oncogenic circuitry by reversing aberrant eIF4F-mediated translation.
Bitterman PB, Polunovsky VA. Bitterman PB, et al. Mol Cancer Ther. 2012 May;11(5):1051-61. doi: 10.1158/1535-7163.MCT-11-0530. Mol Cancer Ther. 2012. PMID: 22572598 Free PMC article. Review.
Cited by
- Translational buffering by ribosome stalling in upstream open reading frames.
Bottorff TA, Park H, Geballe AP, Subramaniam AR. Bottorff TA, et al. PLoS Genet. 2022 Oct 31;18(10):e1010460. doi: 10.1371/journal.pgen.1010460. eCollection 2022 Oct. PLoS Genet. 2022. PMID: 36315596 Free PMC article. - Expansion of human hematopoietic stem cells by inhibiting translation.
Li C, Shin H, Bhavanasi D, Liu M, Yu X, Peslak SA, Liu X, Alvarez-Dominguez JR, Blobel GA, Gregory BD, Huang J, Klein PS. Li C, et al. bioRxiv [Preprint]. 2023 Nov 29:2023.11.28.568925. doi: 10.1101/2023.11.28.568925. bioRxiv. 2023. PMID: 38077058 Free PMC article. Preprint. - RNA-Binding Proteins in Cancer: Functional and Therapeutic Perspectives.
Kang D, Lee Y, Lee JS. Kang D, et al. Cancers (Basel). 2020 Sep 21;12(9):2699. doi: 10.3390/cancers12092699. Cancers (Basel). 2020. PMID: 32967226 Free PMC article. Review. - Translational control in cellular and developmental processes.
Kong J, Lasko P. Kong J, et al. Nat Rev Genet. 2012 Jun;13(6):383-94. doi: 10.1038/nrg3184. Nat Rev Genet. 2012. PMID: 22568971 Review. - eIF4E-Dependent Translational Control: A Central Mechanism for Regulation of Pain Plasticity.
Uttam S, Wong C, Price TJ, Khoutorsky A. Uttam S, et al. Front Genet. 2018 Oct 24;9:470. doi: 10.3389/fgene.2018.00470. eCollection 2018. Front Genet. 2018. PMID: 30459806 Free PMC article. Review.
References
- Gingras AC, Raught B, Sonenberg N. eIF4 initiation factors: effectors of mRNA recruitment to ribosomes and regulators of translation. Annu Rev Biochem. 1999;68:913–963. - PubMed
- Graff JR, Konicek BW, Carter JH, Marcusson EG. Targeting the eukaryotic translation initiation factor 4E for cancer therapy. Cancer Res. 2008;68:631–634. - PubMed
- Guertin DA, Sabatini DM. The pharmacology of mTOR inhibition. Sci Signal. 2009;2:pe24. - PubMed
- Armengol G, et al. 4E-binding protein 1: a key molecular “funnel factor” in human cancer with clinical implications. Cancer Res. 2007;67:7551–7555. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- MH081216-01/MH/NIMH NIH HHS/United States
- R01 GM064700-09/GM/NIGMS NIH HHS/United States
- R01 GM064700/GM/NIGMS NIH HHS/United States
- 1 R01 CA114475/CA/NCI NIH HHS/United States
- R01 CA114475/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous