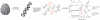Tissue distribution of 5-hydroxymethylcytosine and search for active demethylation intermediates - PubMed (original) (raw)
Tissue distribution of 5-hydroxymethylcytosine and search for active demethylation intermediates
Daniel Globisch et al. PLoS One. 2010.
Abstract
5-Hydroxymethylcytosine (hmC) was recently detected as the sixth base in mammalian tissue at so far controversial levels. The function of the modified base is currently unknown, but it is certain that the base is generated from 5-methylcytosine (mC). This fuels the hypothesis that it represents an intermediate of an active demethylation process, which could involve further oxidation of the hydroxymethyl group to a formyl or carboxyl group followed by either deformylation or decarboxylation. Here, we use an ultra-sensitive and accurate isotope based LC-MS method to precisely determine the levels of hmC in various mouse tissues and we searched for 5-formylcytosine (fC), 5-carboxylcytosine (caC), and 5-hydroxymethyluracil (hmU) as putative active demethylation intermediates. Our data suggest that an active oxidative mC demethylation pathway is unlikely to occur. Additionally, we show using HPLC-MS analysis and immunohistochemistry that hmC is present in all tissues and cell types with highest concentrations in neuronal cells of the CNS.
Conflict of interest statement
Competing Interests: Funding was received from Bayer-Schering Pharma AG, which does not alter the authors' adherence to the policies of PLoS ONE.
Figures
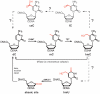
Figure 1. Putative demethylation pathways.
Depiction of the known cytosine modifications mC and hmC and of the putative oxidative “demethylation” intermediates fC and caC. The base excision repair (BER) pathway is a second possible demethylation pathway via the intermediate hmU.
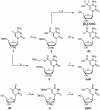
Figure 2. Synthesis of [D2]-hmC and the putative intermediates fC, caC, and hmU.
a) CO, PPh3, Pd2(dba)3·CHCl3, Bu3SnH, 97%, with Bu3SnD 49%, b) HF pyridine, 75%, c) NaBD4, CeCl3·7H2O, 21%, d) TBAF, 51%, e) CO, TMS-Et-OH, DIPEA, Pd2Cl2(MeCN)2, 52%, f) TBAF, 69%, g) Reference h) HF·pyridine, 70%. All reactions could also be carried out in a protective group free manner but resulted in reduced yields and tedious workups (see Figure S2 for details).
Figure 3. Workflow of the HPLC-MS quantification method.
DNA is extracted from any kind of tissue and subsequently enzymatically digested to the nucleosides. Subsequently, a known amount of the stable isotope-labeled standard nucleoside is added. In the HPLC-MS analysis one signal for the natural (light) and one for the synthetic (heavy) compound is detected in each experiment. Quantification is performed by comparing the integrals of the specific high resolution ion current of the natural compound (amount to be determined) with their corresponding heavy atom labeled derivative (known amount).
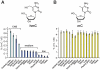
Figure 4. Quantification of hmC and mC in mouse tissue.
A) Values of hmC in % of dG. B) Values of mC in % of dG. A)+B) Data represent values for each tissue of at least two mice with standard deviation (SD) (See Table S1 for details). Light-colored bars represent data from our earlier study .
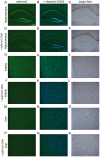
Figure 5. Immunolocalization of hmC in mouse hippocampus, kidney, and liver.
Scale bar: 200 µM. Left column: mouse tissues stained with anti-hmC (green). Middle column: mouse tissues stained with anti-hmC (green) and Hoechst 33342 (blue) for nuclear staining. Right column: Bright field pictures of corresponding tissue. In every second row 2 µM hmC-DNA were added to compete the anti-hmC staining signal out.
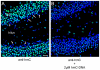
Figure 6. Immunolocalization of hmC in mouse hippocampus.
High magnification images of hmC immunoreactivity in the dentate gyrus (DG) and the hilus of mouse hippocampus. A) Signal for anti-hmC (green) and Hoechst 33342 nuclear dye (blue) B) Competition of anti-hmC with 2 µM hmC-DNA. The scale bar marks 20 µm.
Figure 7. Detection of potential demethylation intermediates caC, hmU, and fC.
A) HPLC-chromatogram of the synthesized cytosine and uracil modifications caC, hmC, hmU, mC, and fC as 2′-deoxynucleosides showing excellent separation of the compounds. B) Detected values of the potential intermediates as example in olfactory bulb. The red line indicates the detection limits of the modified nucleosides in enzymatically digested DNA samples.
Similar articles
- Tissue-Specific Differences in DNA Modifications (5-Hydroxymethylcytosine, 5-Formylcytosine, 5-Carboxylcytosine and 5-Hydroxymethyluracil) and Their Interrelationships.
Gackowski D, Zarakowska E, Starczak M, Modrzejewska M, Olinski R. Gackowski D, et al. PLoS One. 2015 Dec 14;10(12):e0144859. doi: 10.1371/journal.pone.0144859. eCollection 2015. PLoS One. 2015. PMID: 26660343 Free PMC article. - Improved synthesis and mutagenicity of oligonucleotides containing 5-hydroxymethylcytosine, 5-formylcytosine and 5-carboxylcytosine.
Münzel M, Lischke U, Stathis D, Pfaffeneder T, Gnerlich FA, Deiml CA, Koch SC, Karaghiosoff K, Carell T. Münzel M, et al. Chemistry. 2011 Dec 2;17(49):13782-8. doi: 10.1002/chem.201102782. Epub 2011 Nov 8. Chemistry. 2011. PMID: 22069110 - 5-Hydroxymethylcytosine, the sixth base of the genome.
Münzel M, Globisch D, Carell T. Münzel M, et al. Angew Chem Int Ed Engl. 2011 Jul 11;50(29):6460-8. doi: 10.1002/anie.201101547. Epub 2011 Jun 17. Angew Chem Int Ed Engl. 2011. PMID: 21688365 Review. - 5-Hydroxymethylcytosine: the many faces of the sixth base of mammalian DNA.
Kriukienė E, Tomkuvienė M, Klimašauskas S. Kriukienė E, et al. Chem Soc Rev. 2024 Mar 4;53(5):2264-2283. doi: 10.1039/d3cs00858d. Chem Soc Rev. 2024. PMID: 38205583 Review.
Cited by
- DNA Methylation Analysis by Bisulfite Pyrosequencing of Mouse Embryonic Fibroblasts with Reprogramming Enhanced by Thyroid Hormones.
Santamarina-Ojeda P, Fernández AF, Fraga MF, Pérez RF. Santamarina-Ojeda P, et al. Methods Mol Biol. 2025;2876:131-147. doi: 10.1007/978-1-0716-4252-8_9. Methods Mol Biol. 2025. PMID: 39579313 - Bacteriophage-related epigenetic natural and non-natural pyrimidine nucleotides and their influence on transcription with T7 RNA polymerase.
Gracias F, Pohl R, Sýkorová V, Hocek M. Gracias F, et al. Commun Chem. 2024 Nov 9;7(1):256. doi: 10.1038/s42004-024-01354-5. Commun Chem. 2024. PMID: 39521867 Free PMC article. - Loss of TET Activity in the Postnatal Mouse Brain Perturbs Synaptic Gene Expression and Impairs Cognitive Function.
Liu JW, Zhang ZQ, Zhu ZC, Li K, Xu Q, Zhang J, Cheng XW, Li H, Sun Y, Wang JJ, Hu LL, Xiong ZQ, Zhu Y. Liu JW, et al. Neurosci Bull. 2024 Nov;40(11):1699-1712. doi: 10.1007/s12264-024-01302-2. Epub 2024 Oct 12. Neurosci Bull. 2024. PMID: 39395911 - Cell-Free DNA Hydroxymethylation in Cancer: Current and Emerging Detection Methods and Clinical Applications.
Li JJN, Liu G, Lok BH. Li JJN, et al. Genes (Basel). 2024 Sep 3;15(9):1160. doi: 10.3390/genes15091160. Genes (Basel). 2024. PMID: 39336751 Free PMC article. Review. - Implementation of the Methyl-Seq platform to identify tissue- and sex-specific DNA methylation differences in the rat epigenome.
Cox OH, Seifuddin F, Guo J, Pirooznia M, Boersma GJ, Wang J, Tamashiro KLK, Lee RS. Cox OH, et al. Epigenetics. 2024 Dec;19(1):2393945. doi: 10.1080/15592294.2024.2393945. Epub 2024 Sep 22. Epigenetics. 2024. PMID: 39306700 Free PMC article.
References
- Liutkeviciute Z, Lukinavicius G, Masevicius V, Daujotyte D, Klimasauskas S. Cytosine-5-methyltransferases add aldehydes to DNA. Nat Chem Biol. 2009;5:400–402. - PubMed
- Miller CA, Sweatt JD. Covalent modification of DNA regulates memory formation. Neuron. 2007;53:857–869. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources