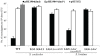Cleavage of phosphorothioated DNA and methylated DNA by the type IV restriction endonuclease ScoMcrA - PubMed (original) (raw)
Cleavage of phosphorothioated DNA and methylated DNA by the type IV restriction endonuclease ScoMcrA
Guang Liu et al. PLoS Genet. 2010.
Abstract
Many taxonomically diverse prokaryotes enzymatically modify their DNA by replacing a non-bridging oxygen with a sulfur atom at specific sequences. The biological implications of this DNA S-modification (phosphorothioation) were unknown. We observed that simultaneous expression of the dndA-E gene cluster from Streptomyces lividans 66, which is responsible for the DNA S-modification, and the putative Streptomyces coelicolor A(3)2 Type IV methyl-dependent restriction endonuclease ScoA3McrA (Sco4631) leads to cell death in the same host. A His-tagged derivative of ScoA3McrA cleaved S-modified DNA and also Dcm-methylated DNA in vitro near the respective modification sites. Double-strand cleavage occurred 16-28 nucleotides away from the phosphorothioate links. DNase I footprinting demonstrated binding of ScoA3McrA to the Dcm methylation site, but no clear binding could be detected at the S-modified site under cleavage conditions. This is the first report of in vitro endonuclease activity of a McrA homologue and also the first demonstration of an enzyme that specifically cleaves S-modified DNA.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
Figure 1. Streptomyces and E. coli strains used as sources of recombinant DNA.
Gi11 ( = SLP1) and SLG are genomic islands (DNA inserts) which are unique to S. coelicolor A(3)2 and S. lividans 66, respectively. The numbers on the arrows between the E. coli and Streptomyces cartoons indicate the approximate relative efficiency of plasmid transfer by protoplast transformation as reported by Gonzalez-Ceron et al. (2009). McrA (ScoA3McrA) is remotely similar to the Type IV restriction endonucleases EcoKMcrA and has been shown to restrict methylated DNA in vivo in Stretpomyces. Dam, Dcm and Hsd are sequence-specific DNA methylases of E. coli. dndA-E is a gene cluster of S. lividans responsible for the sequence-specific S-modification in this strain (only c. 1 in 6000 bp contain sulfur; dnd stands for
DN
A
d
egradation during electrophoresis).
Figure 2. Restriction of plasmids expressing the dndA-E gene cluster by Streptomyces strains containing sco4631.
Graph showing the number of apramycin resistant S. coelicolor (1–3) and S. lividans (4 and 5) exconjugants that were obtained in matings with E. coli ET12567/pUZ8002 containing mobilizable (oriT) plasmids. WT, plasmid-free, wild-type S. coelicolor M145 strain; LG3, S. coelicolor mutant lacking the entire genomic island Gi11 ( = SLP1) which includes sco4631; LG4, S. coelicolor mutant from which only sco4631 was deleted; LG5, S. lividans derivative which lacks the dnd gene cluster and contains a cloned copy of the S. coelicolor gene sco4631; LG6 S. lividans derivative which lacks the dnd gene cluster. The graph shows that pHZ1904 which contains the dndA-E gene cluster (causes DNA S-modification; black bars), is specifically restricted by the wild type S. coelicolor strain. pHZ1904* (white bars), which differs from pHZ1904 by an inactivating frame shift point mutation in dndE, was not restricted. The cloning vector pSET152 without insert (grey bar) served as an additional, not restricted control.
Figure 3. Divalent cation ion requirements analysis for the cleavage of S. lividans 1326 total DNA by Sco4631.
Total S. lividans 1326 DNA (100 ng) prepared by the Kirby mix procedure was used as a substrate to react with 500 ng purified His6-Sco4631 protein at 30°C for 5 min in a total reaction volume of 20 µl. The metal ions added to the reaction buffer are indicated to the left, and the concentrations are above the lanes. The reactions were stopped by adding loading buffer containing 1% SDS, 50% glycerol and 0.05% bromophenol blue (Takara). After the reaction, the DNA samples were examined by 0.75% agarose gel electrophoresis. The gel was run at 5 V/cm for 30 min and visualized by staining with ethidium bromide. The strong bands are high molecular weight DNA; the smear observed with Mn2+ and Co2+ concentrations ≥100 µM indicates DNA cleavage by Sco4631.
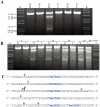
Figure 4. Sco4631-mediated in vitro cleavage of Dcm-methylated plasmid DNA.
A. Agarose gel showing that DNA cleavage by His6-Sco4631 requires DNA methylation. The ethidium bromide-stained agarose gel shows EcoRV linearized pOJ260 DNA treated as indicated above the lanes. Incubation was for 5 min at 30°C in buffer containing 1 mM Mn2+. H, heat inactivated His6-Sco4631 used as a control; E (Enzyme) and E*, His6-Sco4631 and inactive His6-Sco4631(H508A), respectively, purified from E. coli BL21(DE3); K, kb ladder DNA standard; M (methylated), indicates Dcm methylated pOJ260 that was prepared from Dam+ Dcm+ host E. coli DH10B. U (unmethylated), indicates pOJ260 that was isolated from non-methylating E. coli ET12567. The 0.9–2.6 kb bands generated by Sco4631 digestion of methylated DNA are relatively faint, indicating partial digestion. B. Prolonged incubation with His6-Sco4631 causes non-specific DNA degradation, also of unmethylated DNA. EcoRV linearized pOJ260 DNA (like in panel A) was incubated for 5–60 min as indicated above the gel. M, methylated DNA; U, unmethylated DNA. C. Sequences of the cleavage sites including the nearby Dcm methylation sites (C5mCWGG, shown in blue). Vertical arrows point to the cleaved bonds, most cuts were observed once except for the cut marked by a larger arrow that was observed in four independent samples. Note, there are four additional C5mCWGG sequences in pOJ260 (not shown) where no cleavage was observed.
Figure 5. In vitro DNase I protection and cleavage of a double-stranded 164 bp Dcm-methylated synthetic oligonucleotide by purified His6-Sco4631.
A. Analysis of 5′ labeled top strand DNA, and B, 5′ labeled bottom strand DNA. Bands a-d on these autoradiographs represent cleavage of the DNA strands by His6-tagged Sco4631. Lanes 1–3 are controls lacking either the enzyme (E) or the Dcm methylation (M). The samples in lanes 4 contained both methylated DNA and active His6-Sco4631. Lanes 5–8 are sequencing ladders. Lanes 9–12 are controls containing unmethylated DNA and increasing amounts of active His6-Sco4631. Lanes 13–16 contained methylated DNA and increasing amounts of the active His6-Sco4631. Lanes 9 and 13 contained no enzyme, and lanes 10–12 and lanes 14–16 contained 1.1, 4.5 and 18 µM enzyme, respectively. The vertical sequence to the right of each gel picture indicates the DNA regions that were partially protected from cleavage by DNase I. The horizontal lines point to bases that are not protected and shown in lower case letters. (Please note, panel A shows the correct sequence of the top strand, but in panel B there is a compression of the sequence with the bold bases in the sequence CCAGGTGCGAATAAG not visible.) C. Central sequence of the 164 bp double-stranded oligonucleotide containing the Dcm methylated sequence C5mCWGG (dark blue) and additional sequences protected against DNase I activity (light blue). The fat vertical arrows labeled a and b are the major cut sites, and the thin arrows labeled c and d are minor cut sites indicated in the gels A and B. Arrows with numbers indicate cut sites that were identified by cloning and sequencing (see Figure 3). Bases printed light grey are not visible on the gel sections shown.
Figure 6. In vitro cleavage of S-modified (phosphorothioated) DNA by Tris-peracid or His6-Sco4631.
A. Ethidium bromide-stained agarose gel showing EcoRV-linearized pHZ209 DNA (5.4 kb). K, kb ladder DNA size standard; S, S-modified DNA prepared from S. lividans 1326; E, His6-Sco4631; E*, inactive His6-Sco4631(H508A); X, Tris-peracid. Non-phosphorothioated DNA was prepared from S. lividans HXY6. Partial digestion producing strong 3.1 and 2.3 kb bands and additional fainter bands was observed in the samples containing S-modified DNA (S) and either His6-Sco4631 (E), or Tris-peracid. B. Summary of preferential cleavage sites on Dnd-phosphorothioated pHZ209. The two main bands (3.1 kb and 2.3 kb in Figure 5A) from the His6-Sco4631-digested S-modified DNA were cloned into pBluescript SK+, and 16 clones were end-sequenced from both sides using primers T3 and T7. Vertical arrows show where the cuts occurred, and the numbers indicate how many times each particular cut was observed. All 16 cuts occurred near the preferential Dnd-phosphorothioation modification sequence CGpsGCCG shown in blue.
Figure 7. In vitro cleavage of a synthetic S-modified (phosphorothioated) double-stranded 118 bp oligonucleotide by purified His6-Sco4631.
A. Autoradiographs of 5′ labeled top strand DNA (left) and 5′ labeled bottom strand DNA (right). G, A, T, C denote the sequencing ladders. Horizontal arrows indicate cleavage sites, and ** indicates the tandem G residues linked by a phosphorothioate bond. His6-Sco4631-mediated cleavage was observed exclusively in the samples of S-modified DNA (S) that also contained active His6-Sco4631 (E). Samples without DNA S modification or containing heat-inactivated His6-Sco4631 were not cleaved. - indicates the absence of enzyme or S-modification, respectively. B. In vitro cleavage of S-modified DNA by His6-Sco4631. Vertical arrows indicate bonds where cleavage of the 118 bp oligonucleotide was observed. The length of the solid arrows indicates the intensity of the bands in panel A and thus the frequency of cleavage. Dotted arrows indicate in vitro generated cleavage sites of pHZ209 (see Figure 5). The preferred S-modification sequence CGpsGCCG is shown in blue.
Similar articles
- Crystallization and preliminary X-ray analysis of the type IV restriction endonuclease ScoMcrA from Streptomyces coelicolor, which cleaves both Dcm-methylated DNA and phosphorothioated DNA.
Liu G, Zhang Z, Zhao G, Deng Z, Wu G, He X. Liu G, et al. Acta Crystallogr F Struct Biol Commun. 2015 Jan 1;71(Pt 1):57-60. doi: 10.1107/S2053230X14025801. Epub 2015 Jan 1. Acta Crystallogr F Struct Biol Commun. 2015. PMID: 25615970 Free PMC article. - Identification of a conserved DNA sulfur recognition domain by characterizing the phosphorothioate-specific endonuclease SprMcrA from Streptomyces pristinaespiralis.
Yu H, Liu G, Zhao G, Hu W, Wu G, Deng Z, He X. Yu H, et al. Mol Microbiol. 2018 Nov;110(3):484-497. doi: 10.1111/mmi.14118. Epub 2018 Oct 5. Mol Microbiol. 2018. PMID: 30184284 - Characterization of the methyl-specific restriction system of Streptomyces coelicolor A3(2) and of the role played by laterally acquired nucleases.
González-Cerón G, Miranda-Olivares OJ, Servín-González L. González-Cerón G, et al. FEMS Microbiol Lett. 2009 Nov;301(1):35-43. doi: 10.1111/j.1574-6968.2009.01790.x. Epub 2009 Sep 16. FEMS Microbiol Lett. 2009. PMID: 19796133 - Twenty years hunting for sulfur in DNA.
Chen S, Wang L, Deng Z. Chen S, et al. Protein Cell. 2010 Jan;1(1):14-21. doi: 10.1007/s13238-010-0009-y. Epub 2010 Feb 7. Protein Cell. 2010. PMID: 21203994 Free PMC article. Review. - [Research progress and prospects of phosphorothioate modification - A review].
Gao Y, Deng Z, Chen S. Gao Y, et al. Wei Sheng Wu Xue Bao. 2016 Dec 4;56(12):1831-9. Wei Sheng Wu Xue Bao. 2016. PMID: 29741847 Review. Chinese.
Cited by
- Nicking mechanism underlying the DNA phosphorothioate-sensing antiphage defense by SspE.
Gao H, Gong X, Zhou J, Zhang Y, Duan J, Wei Y, Chen L, Deng Z, Wang J, Chen S, Wu G, Wang L. Gao H, et al. Nat Commun. 2022 Nov 9;13(1):6773. doi: 10.1038/s41467-022-34505-0. Nat Commun. 2022. PMID: 36351933 Free PMC article. - Protein Domain Guided Screen for Sequence Specific and Phosphorothioate-Dependent Restriction Endonucleases.
Lutz T, Czapinska H, Fomenkov A, Potapov V, Heiter DF, Cao B, Dedon P, Bochtler M, Xu SY. Lutz T, et al. Front Microbiol. 2020 Aug 18;11:1960. doi: 10.3389/fmicb.2020.01960. eCollection 2020. Front Microbiol. 2020. PMID: 33013736 Free PMC article. - Structural basis for the recognition of sulfur in phosphorothioated DNA.
Liu G, Fu W, Zhang Z, He Y, Yu H, Wang Y, Wang X, Zhao YL, Deng Z, Wu G, He X. Liu G, et al. Nat Commun. 2018 Nov 8;9(1):4689. doi: 10.1038/s41467-018-07093-1. Nat Commun. 2018. PMID: 30409991 Free PMC article. - ICEberg: a web-based resource for integrative and conjugative elements found in Bacteria.
Bi D, Xu Z, Harrison EM, Tai C, Wei Y, He X, Jia S, Deng Z, Rajakumar K, Ou HY. Bi D, et al. Nucleic Acids Res. 2012 Jan;40(Database issue):D621-6. doi: 10.1093/nar/gkr846. Epub 2011 Oct 18. Nucleic Acids Res. 2012. PMID: 22009673 Free PMC article. - Expression and purification of the modification-dependent restriction enzyme BisI and its homologous enzymes.
Xu SY, Klein P, Degtyarev SKh, Roberts RJ. Xu SY, et al. Sci Rep. 2016 Jun 29;6:28579. doi: 10.1038/srep28579. Sci Rep. 2016. PMID: 27353146 Free PMC article.
References
- Robertson KD. DNA Methylation and human disease. Nat Rev Genet. 2005;6:597–610. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases