Initiator elements function to determine the activity state of BX-C enhancers - PubMed (original) (raw)
Initiator elements function to determine the activity state of BX-C enhancers
Carole Iampietro et al. PLoS Genet. 2010.
Abstract
A >300 kb cis-regulatory region is required for the proper expression of the three bithorax complex (BX-C) homeotic genes. Based on genetic and transgenic analysis, a model has been proposed in which the numerous BX-C cis-regulatory elements are spatially restricted through the activation or repression of parasegment-specific chromatin domains. Particular early embryonic enhancers, called initiators, have been proposed to control this complex process. Here, in order to better understand the process of domain activation, we have undertaken a systematic in situ dissection of the iab-6 cis-regulatory domain using a new method, called InSIRT. Using this method, we create and genetically characterize mutations affecting iab-6 function, including mutations specifically modifying the iab-6 initiator. Through our mutagenesis of the iab-6 initiator, we provide strong evidence that initiators function not to directly control homeotic gene expression but rather as domain control centers to determine the activity state of the enhancers and silencers within a cis-regulatory domain.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
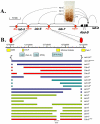
Figure 1. Synopsis of the Abd-B locus of the BX-C and diagram of the mutations created for this study.
A. Synopsis of the Abd-B locus of the BX-C. Diagram of the Abd-B gene and its 3′_cis-_regulatory region. The horizontal line represents the DNA sequence of the BX-C (see scale on top left). The Abd-B expression pattern in the central nervous system of a 10 hours embryo is shown above the DNA line. In parasegment 10 (PS10) Abd-B is present in a few nuclei at a relatively low level. This PS10-specific expression pattern is controlled by the iab-5 regulatory domain located 55 kb downstream from the Abd-B promoter. In PS11, PS12 and PS13, Abd-B is present in progressively more nuclei and at higher levels. These patterns are controlled by the iab-6, iab-7 and iab-8 regulatory domains, respectively. Each regulatory domain functions autonomously from its neighbors due to the presence of the boundaries that flank them (red ovals). B. Diagram of the mutations created for this study. The top line shows the DNA coordinates of iab-6, according to the Drosophila Genome Project. Below this line, and to approximate scale, are the locations of the various elements isolated from the BX-C including the IAB5 initiator, DNase hypersentive site 1 (HS1/Fab-6 including the CTCF binding sites) and 2 (HS2/PRE) –, the 2.8 kb iab-6 initiator fragment , the minimal initiator fragment and the Fab-7 boundary , . Below this line are the DNAs reintegrated to make the mutations. The various iab-6 alleles are indicated as solid bars, with gaps indicating the areas deleted. These bars are color coded such that blue bars indicate mutants that show no cuticle or CNS phenotypes at 25°C, red bars indicate mutants with _Fab-6_-type phenotypes, turquoise bars indicate mutants with iab-5,6 phenotypes, and green bars indicate mutants with iab-6 phenotypes.
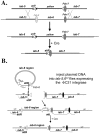
Figure 2. InSIRT.
A. Step one: Homologous Recombination. The original “ends-out” donor vector (pW25) was modified to contain an attP insertion site and a removable yellow reporter gene. Using the yellow reporter, homologous recombination events could be identified by screening for flies with yellow expression in the A5 and A6 segments (a consequence of having yellow inserted in the iab-5 domain). The yellow reporter could then be removed to leave only the attP site and a single loxP recombination site (white triangle) in place of iab-6. B. Step two: Reintegration. Plasmids containing a 288 bp attB site, a single loxP site, a yellow reporter and a version of the 19.3 kb fragment were injected into iab-5,6CI embryos expressing a maternally supplied φC31 integrase . Integration events were isolated based on yellow gene expression, then crossed to the Cre recombinase to remove the yellow gene and all vector backbone sequence.
Figure 3. iab-5,6CI phenotype and rescue.
A. A wild-type adult male cuticle with A4-A6 labeled. Segment A5 differs from A6 based on the sternite shape and the bristles present on the A5 sternite. For reference, the A6 tergite is indicated by a red arrowhead and the A6 sternite is indicated by a red arrow. B. A wild-type embryonic nerve cord (anterior towards the top) stained with an antibody to Abd-B (brown). Notice the step gradient of Abd-B expression increasing in each parasegment towards the posterior. C. An adult male cuticle of a fly homozygous for the iab-5,6CI chromosome with A5 and A6 transformed towards A4 (notice the A4-like pigmentation on the tergites and the bristled sternites). D. The embryonic nerve cord of homozygous iab-5,6CI embryos shows only a transformation of A6 into A5, as seen by the repetition of PS10/A5-like Abd-B levels in PS11/A6, indicating that the inactivation of iab-5 is incomplete and not seen in the embryo. E. An adult male cuticle from a fly homozygous for the iab-5,6rescue chromosome, where the entire 19.3 kb area deleted in iab-5,6CI is reintegrated into iab-5,6CI, looks completely wild type. F. The complete rescue is confirmed by the wild-type pattern of Abd-B in the embryonic ventral nerve cord.
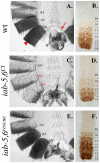
Figure 4. Phenotypes from initiator mutants.
Genotypes are as follows: A.–C. iab-61, D.–F. iab-64 and G.–I. iab-68. A., D. and G. show adult male cuticles. B., E. and H. show pseudo-darkfield views of the fifth and sixth tergites to visualize the trichome patterns. C., F. and I. show the Abd-B staining pattern in the embryonic nerve cord. In wild-type flies, A5/PS10 differs from A6/PS11 based on the sternite shape, the bristles present on the A5 sternite, the trichome pattern on the fifth and sixth tergites, and the Abd-B staining pattern in the CNS (see Figure 3 and Figure 4). The iab-61 and iab-64 show transformations of A6 to A5 for all phenotypes monitored. Meanwhile iab-68 shows only a partial transformation of A6 to A5 as seen by the sternite shape and trichome pattern on A6, which remain A6-like.
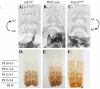
Figure 5. Phenotypes from initiator mutants.
Genotypes are as follows: A. and D. iab-64. B. and E. wild type. C. and F. Fab-6IAB5. A.–C. Show the ventral sternite cuticles made from adult males, homozygous for the genotype indicated above. Notice that A5 differs from A6 based on the sternite shape and the bristles present on the A5 sternite. The opposite homeotic transformations are highlighted by the direction of the arrows on the left and the right of the cuticles. D.–F. Show ventral nerve chords made from homozygous embryos of the genotypes indicated above. Parasegment borders are marked to the left.
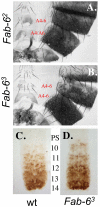
Figure 6. Fab-6 boundary mutations.
The genotypes of the adult male cuticles of A. Fab-62, and B. Fab-63. C. (wild type) and D. (Fab-63) are embryonic nerve cords stained for Abd-B protein. Notice the increased level of Abd-B in PS10 in mutants (D.) relative to wild-type (C.).
Similar articles
- Dissecting the regulatory landscape of the Abd-B gene of the bithorax complex.
Mihaly J, Barges S, Sipos L, Maeda R, Cléard F, Hogga I, Bender W, Gyurkovics H, Karch F. Mihaly J, et al. Development. 2006 Aug;133(15):2983-93. doi: 10.1242/dev.02451. Epub 2006 Jul 3. Development. 2006. PMID: 16818450 - The Fab-8 boundary defines the distal limit of the bithorax complex iab-7 domain and insulates iab-7 from initiation elements and a PRE in the adjacent iab-8 domain.
Barges S, Mihaly J, Galloni M, Hagstrom K, Müller M, Shanower G, Schedl P, Gyurkovics H, Karch F. Barges S, et al. Development. 2000 Feb;127(4):779-90. doi: 10.1242/dev.127.4.779. Development. 2000. PMID: 10648236 - In situ dissection of the Fab-7 region of the bithorax complex into a chromatin domain boundary and a Polycomb-response element.
Mihaly J, Hogga I, Gausz J, Gyurkovics H, Karch F. Mihaly J, et al. Development. 1997 May;124(9):1809-20. doi: 10.1242/dev.124.9.1809. Development. 1997. PMID: 9165128 - Unraveling cis-regulatory mechanisms at the abdominal-A and Abdominal-B genes in the Drosophila bithorax complex.
Akbari OS, Bousum A, Bae E, Drewell RA. Akbari OS, et al. Dev Biol. 2006 May 15;293(2):294-304. doi: 10.1016/j.ydbio.2006.02.015. Epub 2006 Mar 20. Dev Biol. 2006. PMID: 16545794 Review. - Chromatin insulators and long-distance interactions in Drosophila.
Kyrchanova O, Georgiev P. Kyrchanova O, et al. FEBS Lett. 2014 Jan 3;588(1):8-14. doi: 10.1016/j.febslet.2013.10.039. Epub 2013 Nov 5. FEBS Lett. 2014. PMID: 24211836 Review.
Cited by
- Successive gain of insulator proteins in arthropod evolution.
Heger P, George R, Wiehe T. Heger P, et al. Evolution. 2013 Oct;67(10):2945-56. doi: 10.1111/evo.12155. Epub 2013 Jun 4. Evolution. 2013. PMID: 24094345 Free PMC article. - A novel function for the Hox gene Abd-B in the male accessory gland regulates the long-term female post-mating response in Drosophila.
Gligorov D, Sitnik JL, Maeda RK, Wolfner MF, Karch F. Gligorov D, et al. PLoS Genet. 2013 Mar;9(3):e1003395. doi: 10.1371/journal.pgen.1003395. Epub 2013 Mar 28. PLoS Genet. 2013. PMID: 23555301 Free PMC article. - Mechanism and functional role of the interaction between CP190 and the architectural protein Pita in Drosophila melanogaster.
Sabirov M, Kyrchanova O, Pokholkova GV, Bonchuk A, Klimenko N, Belova E, Zhimulev IF, Maksimenko O, Georgiev P. Sabirov M, et al. Epigenetics Chromatin. 2021 Mar 22;14(1):16. doi: 10.1186/s13072-021-00391-x. Epigenetics Chromatin. 2021. PMID: 33752739 Free PMC article. - Homeotic Genes: Clustering, Modularity, and Diversity.
Hajirnis N, Mishra RK. Hajirnis N, et al. Front Cell Dev Biol. 2021 Aug 11;9:718308. doi: 10.3389/fcell.2021.718308. eCollection 2021. Front Cell Dev Biol. 2021. PMID: 34458272 Free PMC article. Review. - Boundary bypass activity in the abdominal-B region of the Drosophila bithorax complex is position dependent and regulated.
Kyrchanova O, Ibragimov A, Postika N, Georgiev P, Schedl P. Kyrchanova O, et al. Open Biol. 2023 Aug;13(8):230035. doi: 10.1098/rsob.230035. Epub 2023 Aug 16. Open Biol. 2023. PMID: 37582404 Free PMC article.
References
- Lewis EB. A gene complex controlling segmentation in Drosophila. Nature. 1978;276:565–570. - PubMed
- Sanchez-Herrero E, Vernos I, Marco R, Morata G. Genetic organization of Drosophila bithorax complex. Nature. 1985;313:108–113. - PubMed
- Maeda RK, Karch F. The ABC of the BX-C: the bithorax complex explained. Development. 2006;133:1413–1422. - PubMed
- Peifer M, Karch F, Bender W. The bithorax complex: control of segmental identity. Genes Dev. 1987;1:891–898. - PubMed
- Sanchez-Herrero E. Control of the expression of the bithorax complex genes abdominal-A and abdominal-B by cis-regulatory regions in Drosophila embryos. Development. 1991;111:437–449. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases