Origin and post-glacial dispersal of mitochondrial DNA haplogroups C and D in northern Asia - PubMed (original) (raw)
Origin and post-glacial dispersal of mitochondrial DNA haplogroups C and D in northern Asia
Miroslava Derenko et al. PLoS One. 2010.
Abstract
More than a half of the northern Asian pool of human mitochondrial DNA (mtDNA) is fragmented into a number of subclades of haplogroups C and D, two of the most frequent haplogroups throughout northern, eastern, central Asia and America. While there has been considerable recent progress in studying mitochondrial variation in eastern Asia and America at the complete genome resolution, little comparable data is available for regions such as southern Siberia--the area where most of northern Asian haplogroups, including C and D, likely diversified. This gap in our knowledge causes a serious barrier for progress in understanding the demographic pre-history of northern Eurasia in general. Here we describe the phylogeography of haplogroups C and D in the populations of northern and eastern Asia. We have analyzed 770 samples from haplogroups C and D (174 and 596, respectively) at high resolution, including 182 novel complete mtDNA sequences representing haplogroups C and D (83 and 99, respectively). The present-day variation of haplogroups C and D suggests that these mtDNA clades expanded before the Last Glacial Maximum (LGM), with their oldest lineages being present in the eastern Asia. Unlike in eastern Asia, most of the northern Asian variants of haplogroups C and D began the expansion after the LGM, thus pointing to post-glacial re-colonization of northern Asia. Our results show that both haplogroups were involved in migrations, from eastern Asia and southern Siberia to eastern and northeastern Europe, likely during the middle Holocene.
Conflict of interest statement
Competing Interests: The authors have declared that no competing interests exist.
Figures
Figure 1. Complete mtDNA phylogenetic tree of haplogroup C.
This schematic tree is based on phylogenetic tree presented in Figure S1. Time estimates (in kya) shown for mtDNA subclusters are based on the complete mtDNA genome clock (the first value) and the synonymous clock (the second value) . The size of each circle is proportional to the number of individuals sharing the corresponding haplotype, with the smallest size corresponding to one individual. Geographic origin is indicated by different colors: northeastern Asian – in blue, central and southern Siberian – in green, eastern Asian – in red, Indian – in grey, European – in yellow, and others (i.e. of unknown population origin) – in white.
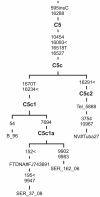
Figure 2. Complete mtDNA phylogenetic tree of subhaplogroup C5c.
Numbers along links refer to substitutions scored relative to rCRS . Transversions are further specified; ins denotes insertion of nucleotide; symbol < denotes parallel mutation. Subject origins are: Poles (B_96; Ser_37_08; Ser_162_06), Teleut (Tel), Tubalar (NV#Tuba from Volodko et al. [12]), and FamilyTreeDNA Project individual of unknown population origin (FTDNA).
Figure 3. Complete mtDNA phylogenetic tree of haplogroup D.
This schematic tree is based on phylogenetic tree presented in Figure S2. Time estimates (in kya) shown for mtDNA subclusters are based on the complete mtDNA genome clock (the first value) and the synonymous clock (the second value) . The size of each circle is proportional to the number of individuals sharing the corresponding haplotype, with the smallest size corresponding to one individual. Geographic origin is indicated by different colors: northeastern Asian – in blue, central and southern Siberian – in green, eastern Asian – in red, Indian – in grey, European – in yellow, and others (i.e. of unknown population origin) – in white.
Figure 4. Complete mtDNA phylogenetic tree of subhaplogroup D4b1a2.
Numbers along links refer to substitutions scored relative to rCRS . Transversions are further specified; symbol < denotes parallel mutation, back mutation is underlined. Subject origins are: Russian (Rus), Buryat (Br), Barghut (Bt), Altaian (Alt), Khamnigan (Hm), Tuvinian (NV#Tuv from Volodko et al. , Yukaghir (NV#Yuk from Volodko et al. [12]), Tubalar (NV#Tub from Volodko et al. [12]), Eskimo (NV#Esk from Volodko et al. [12]), Canadian Inuit (IG#CanInnuit from Ingman, Gyllensten [32]), and Tatar (BM#Tm from Malyarchuk et al. [58]).
Similar articles
- Complete mitochondrial DNA analysis of eastern Eurasian haplogroups rarely found in populations of northern Asia and eastern Europe.
Derenko M, Malyarchuk B, Denisova G, Perkova M, Rogalla U, Grzybowski T, Khusnutdinova E, Dambueva I, Zakharov I. Derenko M, et al. PLoS One. 2012;7(2):e32179. doi: 10.1371/journal.pone.0032179. Epub 2012 Feb 21. PLoS One. 2012. PMID: 22363811 Free PMC article. - Origin and expansion of haplogroup H, the dominant human mitochondrial DNA lineage in West Eurasia: the Near Eastern and Caucasian perspective.
Roostalu U, Kutuev I, Loogväli EL, Metspalu E, Tambets K, Reidla M, Khusnutdinova EK, Usanga E, Kivisild T, Villems R. Roostalu U, et al. Mol Biol Evol. 2007 Feb;24(2):436-48. doi: 10.1093/molbev/msl173. Epub 2006 Nov 10. Mol Biol Evol. 2007. PMID: 17099056 - Phylogeographic analysis of mitochondrial DNA in northern Asian populations.
Derenko M, Malyarchuk B, Grzybowski T, Denisova G, Dambueva I, Perkova M, Dorzhu C, Luzina F, Lee HK, Vanecek T, Villems R, Zakharov I. Derenko M, et al. Am J Hum Genet. 2007 Nov;81(5):1025-41. doi: 10.1086/522933. Epub 2007 Oct 1. Am J Hum Genet. 2007. PMID: 17924343 Free PMC article. - The human genetic history of East Asia: weaving a complex tapestry.
Stoneking M, Delfin F. Stoneking M, et al. Curr Biol. 2010 Feb 23;20(4):R188-93. doi: 10.1016/j.cub.2009.11.052. Curr Biol. 2010. PMID: 20178766 Review. - Human evolutionary history in Eastern Eurasia using insights from ancient DNA.
Zhang M, Fu Q. Zhang M, et al. Curr Opin Genet Dev. 2020 Jun;62:78-84. doi: 10.1016/j.gde.2020.06.009. Epub 2020 Jul 17. Curr Opin Genet Dev. 2020. PMID: 32688244 Review.
Cited by
- Phylogenetic and population genetic analysis of Arunachali Yak (Bos grunniens) using mitochondrial DNA D-loop region.
Pukhrambam M, Dutta A, Das PJ, Chanda A, Sarkar M. Pukhrambam M, et al. Mol Biol Rep. 2024 Nov 6;51(1):1125. doi: 10.1007/s11033-024-10076-9. Mol Biol Rep. 2024. PMID: 39503949 - High prevalence of m.1555A > G in patients with hearing loss in the Baikal Lake region of Russia as a result of founder effect.
Borisova TV, Cherdonova AM, Pshennikova VG, Teryutin FM, Morozov IV, Bondar AA, Baturina OA, Kabilov MR, Romanov GP, Solovyev AV, Fedorova SA, Barashkov NA. Borisova TV, et al. Sci Rep. 2024 Jul 3;14(1):15342. doi: 10.1038/s41598-024-66254-z. Sci Rep. 2024. PMID: 38961196 Free PMC article. - Gene pool preservation across time and space In Mongolian-speaking Oirats.
Balinova N, Hudjašov G, Pankratov V, Pennarun E, Reidla M, Metspalu E, Batyrov V, Khomyakova I, Reisberg T, Parik J, Dzhaubermezov M, Aiyzhy E, Balinova A, El'chinova G, Spitsyna N, Khusnutdinova E, Metspalu M, Tambets K, Villems R, Kushniarevich A. Balinova N, et al. Eur J Hum Genet. 2024 Sep;32(9):1150-1158. doi: 10.1038/s41431-024-01588-w. Epub 2024 Apr 11. Eur J Hum Genet. 2024. PMID: 38605123 Free PMC article. - Whole mitochondrial genome analysis in highland Tibetans: further matrilineal genetic structure exploration.
Li X, Zhang X, Yu T, Ye L, Huang T, Chen Y, Liu S, Wen Y. Li X, et al. Front Genet. 2023 Nov 14;14:1221388. doi: 10.3389/fgene.2023.1221388. eCollection 2023. Front Genet. 2023. PMID: 38034496 Free PMC article. - Unraveling the mitochondrial phylogenetic landscape of Thailand reveals complex admixture and demographic dynamics.
Jaisamut K, Pitiwararom R, Sukawutthiya P, Sathirapatya T, Noh H, Worrapitirungsi W, Vongpaisarnsin K. Jaisamut K, et al. Sci Rep. 2023 Nov 21;13(1):20396. doi: 10.1038/s41598-023-47762-w. Sci Rep. 2023. PMID: 37990137 Free PMC article.
References
- Pitulko VV, Nikolsky PA, Girya EY, Basilyan AE, Tumskoy VE, et al. The Yana RHS site: humans in the Arctic before the last glacial maximum. Science. 2004;303:52–56. - PubMed
- Goebel T, Waters MR, O'Rourke DH. The late Pleistocene dispersal of modern humans in the Americas. Science. 2008;319:1497–1502. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous