Genomic DNA sequences from mastodon and woolly mammoth reveal deep speciation of forest and savanna elephants - PubMed (original) (raw)
Genomic DNA sequences from mastodon and woolly mammoth reveal deep speciation of forest and savanna elephants
Nadin Rohland et al. PLoS Biol. 2010.
Abstract
To elucidate the history of living and extinct elephantids, we generated 39,763 bp of aligned nuclear DNA sequence across 375 loci for African savanna elephant, African forest elephant, Asian elephant, the extinct American mastodon, and the woolly mammoth. Our data establish that the Asian elephant is the closest living relative of the extinct mammoth in the nuclear genome, extending previous findings from mitochondrial DNA analyses. We also find that savanna and forest elephants, which some have argued are the same species, are as or more divergent in the nuclear genome as mammoths and Asian elephants, which are considered to be distinct genera, thus resolving a long-standing debate about the appropriate taxonomic classification of the African elephants. Finally, we document a much larger effective population size in forest elephants compared with the other elephantid taxa, likely reflecting species differences in ancient geographic structure and range and differences in life history traits such as variance in male reproductive success.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
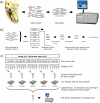
Figure 1. Strategy for obtaining overlapping DNA from four elephantids and a mastodon.
(a) Mastodon shotgun 454 sequencing. We ligated 454-adaptors (green and blue) to the ends of the DNA molecules and sequenced the libraries on a Roche 454 GS. (b) Bioinformatic analysis of shotgun 454 sequences. To identify proboscidean sequence, we compared the sequences to databases consisting of the savanna elephant draft genome (loxAfr1), the human genome (hg18), the mouse genome (mm8), NCBI's nucleotide database of environmental samples (env), and NCBI's non-redundant nucleotide database (nr). The 454 sequences with a best match to loxAfr1 (in red) were aligned to loxAfr1. Alignments of at least 90 bp in length and with a similarity higher than 87% were used for primer design after filtering out known repeat elements (using the UCSC RepeatMasker database). Primers were based on loxAfr1 sequence flanking the mastodon sequence. (c) Multiplex PCR and sequencing of the targeted loci in modern elephants and mammoth. We show the protocol for the first of four rounds of the project (Table S3 provides details of the further rounds). A total of 213 primer pairs were randomly divided into 5 multiplex primer mixes with 41–44 primer pairs per mix. These mixes were used for the first step of the two-step multiplex PCR approach, for each of the 5 samples (La, Loxodonta africana; Lc, L. cyclotis; Em 1, Elephas maximus 1; Em 2, E. maximus 2; Mp, Mammuthus primigenius). Dilutions of these products were used as templates to amplify the loci individually in the second step (shown for L. africana), resulting in 213 distinct products per sample. These products were quantified, normalized, and merged into one pool per sample. A 454 library was prepared and sequenced on 1/16th of a picotiter plate of a Roche 454 GS.
Figure 2. Demographic model for the history of the Elephantidae.
Demographic model that is fit by MCMCcoal, in which all population splits are instantaneous (without subsequent gene flow), and all population sizes are assumed to be constant over intervals. Here, T FS refers to forest-savanna elephant population divergence time, T AM refers to Asian elephant-mammoth population divergence time, T Lox-Eur refers to African-Eurasian population divergence time, and T Elephantid-Mastodon refers to elephantid-mastodon population divergence time, presented here in millions of years. The Ν quantities refer to constant diploid effective population sizes ancestral to each of these splits (in thousands). For obtaining estimates of years and population sizes, we assume that the elephantids have an average of 31 years per generation, based on estimates of 17–20 years for females , and 40–49 years for males ,. A lower or higher number of years per generation would produce a proportionate effect on the population size estimates. For each parameter, two sets of numbers are shown. The upper set shows the range consistent with the fossil record, calibrating to an assumed African-Eurasian population split of T Lox-Eur = 4.2–9 Mya (justified in Text S5). For example for forest-savanna population divergence, this leads to T FS = 2.6–5.6 Mya given that MCMCcoal estimates T FS /T Lox-Eur = 62%. The lower set of numbers (in parentheses) provides MCMCcoal's 90% credible interval for the parameters as a fraction of the best estimate (e.g. 76%–126% for T FS). In the main text, we conservatively quote a range that combines the uncertainty from the fossil record and from MCMCcoal (e.g. T FS = 1.9–7.1 Mya).
Similar articles
- A comprehensive genomic history of extinct and living elephants.
Palkopoulou E, Lipson M, Mallick S, Nielsen S, Rohland N, Baleka S, Karpinski E, Ivancevic AM, To TH, Kortschak RD, Raison JM, Qu Z, Chin TJ, Alt KW, Claesson S, Dalén L, MacPhee RDE, Meller H, Roca AL, Ryder OA, Heiman D, Young S, Breen M, Williams C, Aken BL, Ruffier M, Karlsson E, Johnson J, Di Palma F, Alfoldi J, Adelson DL, Mailund T, Munch K, Lindblad-Toh K, Hofreiter M, Poinar H, Reich D. Palkopoulou E, et al. Proc Natl Acad Sci U S A. 2018 Mar 13;115(11):E2566-E2574. doi: 10.1073/pnas.1720554115. Epub 2018 Feb 26. Proc Natl Acad Sci U S A. 2018. PMID: 29483247 Free PMC article. - Forest elephant mitochondrial genomes reveal that elephantid diversification in Africa tracked climate transitions.
Brandt AL, Ishida Y, Georgiadis NJ, Roca AL. Brandt AL, et al. Mol Ecol. 2012 Mar;21(5):1175-89. doi: 10.1111/j.1365-294X.2012.05461.x. Epub 2012 Jan 19. Mol Ecol. 2012. PMID: 22260276 - Elephant natural history: a genomic perspective.
Roca AL, Ishida Y, Brandt AL, Benjamin NR, Zhao K, Georgiadis NJ. Roca AL, et al. Annu Rev Anim Biosci. 2015;3:139-67. doi: 10.1146/annurev-animal-022114-110838. Epub 2014 Dec 8. Annu Rev Anim Biosci. 2015. PMID: 25493538 Review. - Sequencing the nuclear genome of the extinct woolly mammoth.
Miller W, Drautz DI, Ratan A, Pusey B, Qi J, Lesk AM, Tomsho LP, Packard MD, Zhao F, Sher A, Tikhonov A, Raney B, Patterson N, Lindblad-Toh K, Lander ES, Knight JR, Irzyk GP, Fredrikson KM, Harkins TT, Sheridan S, Pringle T, Schuster SC. Miller W, et al. Nature. 2008 Nov 20;456(7220):387-90. doi: 10.1038/nature07446. Nature. 2008. PMID: 19020620 - The mastodon mitochondrial genome: a mammoth accomplishment.
Roca AL. Roca AL. Trends Genet. 2008 Feb;24(2):49-52. doi: 10.1016/j.tig.2007.11.005. Epub 2008 Jan 14. Trends Genet. 2008. PMID: 18192067 Review.
Cited by
- Population genetic structure and historical demography of the population of forest elephants in Côte d'Ivoire.
Kouakou JL, Gonedelé-Bi S. Kouakou JL, et al. PLoS One. 2024 Aug 26;19(8):e0300468. doi: 10.1371/journal.pone.0300468. eCollection 2024. PLoS One. 2024. PMID: 39186735 Free PMC article. - Ancient mitogenomes from Pre-Pottery Neolithic Central Anatolia and the effects of a Late Neolithic bottleneck in sheep (Ovis aries).
Sandoval-Castellanos E, Hare AJ, Lin AT, Dimopoulos EA, Daly KG, Geiger S, Mullin VE, Wiechmann I, Mattiangeli V, Lühken G, Zinovieva NA, Zidarov P, Çakırlar C, Stoddart S, Orton D, Bulatović J, Mashkour M, Sauer EW, Horwitz LK, Horejs B, Atici L, Özkaya V, Mullville J, Parker Pearson M, Mainland I, Card N, Brown L, Sharples N, Griffiths D, Allen D, Arbuckle B, Abell JT, Duru G, Mentzer SM, Munro ND, Uzdurum M, Gülçur S, Buitenhuis H, Gladyr E, Stiner MC, Pöllath N, Özbaşaran M, Krebs S, Burger J, Frantz L, Medugorac I, Bradley DG, Peters J. Sandoval-Castellanos E, et al. Sci Adv. 2024 Apr 12;10(15):eadj0954. doi: 10.1126/sciadv.adj0954. Epub 2024 Apr 12. Sci Adv. 2024. PMID: 38608027 Free PMC article. - African bushpigs exhibit porous species boundaries and appeared in Madagascar concurrently with human arrival.
Balboa RF, Bertola LD, Brüniche-Olsen A, Rasmussen MS, Liu X, Besnard G, Salmona J, Santander CG, He S, Zinner D, Pedrono M, Muwanika V, Masembe C, Schubert M, Kuja J, Quinn L, Garcia-Erill G, Stæger FF, Rakotoarivony R, Henrique M, Lin L, Wang X, Heaton MP, Smith TPL, Hanghøj K, Sinding MS, Atickem A, Chikhi L, Roos C, Gaubert P, Siegismund HR, Moltke I, Albrechtsen A, Heller R. Balboa RF, et al. Nat Commun. 2024 Jan 3;15(1):172. doi: 10.1038/s41467-023-44105-1. Nat Commun. 2024. PMID: 38172616 Free PMC article. - Genetic features of Sri Lankan elephant, Elephas maximus maximus Linnaeus revealed by high throughput sequencing of mitogenome and ddRAD-seq.
Sooriyabandara MGC, Jayasundara JMSM, Marasinghe MSLRP, Hathurusinghe HABM, Bandaranayake AU, Jayawardane KANC, Nilanthi RMR, Rajapakse RC, Bandaranayake PCG. Sooriyabandara MGC, et al. PLoS One. 2023 Jun 13;18(6):e0285572. doi: 10.1371/journal.pone.0285572. eCollection 2023. PLoS One. 2023. PMID: 37310948 Free PMC article. - Shape variation in the limb long bones of modern elephants reveals adaptations to body mass and habitat.
Bader C, Delapré A, Houssaye A. Bader C, et al. J Anat. 2023 May;242(5):806-830. doi: 10.1111/joa.13827. Epub 2023 Feb 23. J Anat. 2023. PMID: 36824051 Free PMC article.
References
- Barnes I, Shapiro B, Lister A, Kuznetsova T, Sher A, et al. Genetic structure and extinction of the woolly mammoth, Mammuthus primigenius. Curr Biol. 2007;17:1072–1075. - PubMed
- Debruyne R, Chu G, King C. E, Bos K, Kuch M, et al. Out of America: ancient DNA evidence for a new world origin of late quaternary woolly mammoths. Curr Biol. 2008;18:1320–1326. - PubMed
- Gilbert M. T, Tomsho L. P, Rendulic S, Packard M, Drautz D. I, et al. Whole-genome shotgun sequencing of mitochondria from ancient hair shafts. Science. 2007;317:1927–1930. - PubMed
- Krause J, Dear P. H, Pollack J. L, Slatkin M, Spriggs H, et al. Multiplex amplification of the mammoth mitochondrial genome and the evolution of Elephantidae. Nature. 2006;439:724–727. - PubMed
- Miller W, Drautz D. I, Ratan A, Pusey B, Qi J, et al. Sequencing the nuclear genome of the extinct woolly mammoth. Nature. 2008;456:387–390. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources