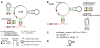R2R--software to speed the depiction of aesthetic consensus RNA secondary structures - PubMed (original) (raw)
R2R--software to speed the depiction of aesthetic consensus RNA secondary structures
Zasha Weinberg et al. BMC Bioinformatics. 2011.
Abstract
Background: With continuing identification of novel structured noncoding RNAs, there is an increasing need to create schematic diagrams showing the consensus features of these molecules. RNA structural diagrams are typically made either with general-purpose drawing programs like Adobe Illustrator, or with automated or interactive programs specific to RNA. Unfortunately, the use of applications like Illustrator is extremely time consuming, while existing RNA-specific programs produce figures that are useful, but usually not of the same aesthetic quality as those produced at great cost in Illustrator. Additionally, most existing RNA-specific applications are designed for drawing single RNA molecules, not consensus diagrams.
Results: We created R2R, a computer program that facilitates the generation of aesthetic and readable drawings of RNA consensus diagrams in a fraction of the time required with general-purpose drawing programs. Since the inference of a consensus RNA structure typically requires a multiple-sequence alignment, the R2R user annotates the alignment with commands directing the layout and annotation of the RNA. R2R creates SVG or PDF output that can be imported into Adobe Illustrator, Inkscape or CorelDRAW. R2R can be used to create consensus sequence and secondary structure models for novel RNA structures or to revise models when new representatives for known RNA classes become available. Although R2R does not currently have a graphical user interface, it has proven useful in our efforts to create 100 schematic models of distinct noncoding RNA classes.
Conclusions: R2R makes it possible to obtain high-quality drawings of the consensus sequence and structural models of many diverse RNA structures with a more practical amount of effort. R2R software is available at http://breaker.research.yale.edu/R2R and as an Additional file.
Figures
Figure 1
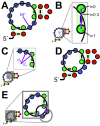
Example of a consensus diagram for a noncoding RNA. (A) Completed diagram of the consensus for the crcB motif [19] created using R2R and Adobe Illustrator. The consensus diagram shown here is modelled on a previously published figure [19]. The legend inset also applies to other consensus diagrams in this report. A generic legend is available for R2R users (Additional files 3 and 4). (B) Raw output generated by R2R when run on the crcB structure. The pseudoknot is depicted separately, along with the hairpin that is present in 31% of crcB RNAs. (C) R2R commands (Additional file 2) used for the main structure in part B. The symbols j, <, >, 1 and 2 in these commands refer to columns in the alignment (explained in Figure 2). (D) Raw output of R2R "skeleton" drawing of the crcB motif.
Figure 2
Complete example using a tiny, contrived RNA. (A) Alignment of fictional RNAs, in Stockholm format [15]. The "#=GC SS_cons" line specifies a stem (shaded blue rectangles) based on matching angle brackets (< and >). The "#=GC R2R_LABEL" line associates the labels [, ], 1, 2 and T with specific columns. The labels are used in R2R markup (e.g., see text "label & use"). (B) Raw output of R2R when run on the input in part A.
Figure 3
Automated layout of multistem junctions. Raw R2R output is shown for the manA motif [19], a pseudoknot (lower right) and a "skeleton"-style drawing (upper right). The layout of the central multistem junction was determined using R2R's solver functions. This motif presents an atypical case where it is not possible to direct all stems horizontally or vertically. The directions of the stems were chosen manually to promote symmetry, the lowest two stems on the junction were constrained to be aligned vertically, and the midpoint of these two stems was aligned horizontally with the upper-most stem. Subject to these constraints, R2R's solver chose a layout that best approximated a circle. The layout was used in a previously published figure [19]. A finished drawing would require assembling the pseudoknot (as in Figure 1), and moving text. Modular structures present in some manA RNAs are not shown. Note: the text "0-6 nt" was moved inward manually to fit the column width.
Figure 4
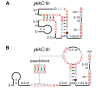
Pseudoknot depiction styles. The _ykkC_-III motif [19] is used to illustrate two styles of drawing pseudoknots (A) "In-line" style. (B) "Callout" style. A portion of this figure was adapted from a previous report [19].
Figure 5
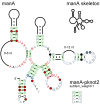
Modular structures in tRNAs and psaA RNAs. (A) Consensus diagram of tRNAs taken from the Rfam database [29] and drawn using the standard tRNA layout. A hairpin that is only sometimes present is shown (lower, right). In this case, the hairpin does not conserve obvious features, and is therefore shown in the "skeleton" style. Other consensus features of tRNAs are not depicted here. (B) Consensus diagram of psaA RNAs [19]. The terminal loops of this RNA often adopt the UNCG tetraloop [21], but also often conform to the CUNG tetraloop [30] or an unstudied CYYGN pentaloop pattern. These distinct sequence features are drawn as modular structures, and were manually positioned near to their associated terminal loop. Other than this repositioning, the diagram is raw R2R output. Some additional annotation and sequence of the psaA motif is not shown here. A portion of this figure was adapted from a previous report [19].
Figure 6
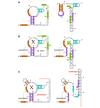
Alternate structures of crcB RNAs. (A) Output of R2R for predicted alternate structures for a crcB RNA in Acidothermus cellulolyticus 11B. Stems are shaded so that their positions in the alternate structures are apparent. R2R commands were used to shade selected nucleotides, to position the multistem junction using the automated solver and to turn the direction of the backbone in two places within the 3' tail. (B) Finished drawing, assembled using Adobe Illustrator based on part A. The predicted Shine-Dalgarno (SD) sequence and start codon are shaded green and labelled. It is hypothesized that when the RNA binds its ligand "X" (left), the SD sequence is available for ribosome binding, allowing gene expression. In the absence of ligand (right), the SD sequence is sequestered, inhibiting gene expression. This latter drawing was made by combining the two drawings in part A. (C) Alternate hypothetical structures of a crcB RNA in Roseburia intestinalis L1-82, finished drawing. The hypothesized structure without the ligand X (right) allows the formation of a putative transcription terminator, which inhibits gene expression. The terminator stem is labelled, and its characteristic poly-U stretch is colored red.
Figure 7
Multistem-junction layout as a non-linear program. (A) Illustration of first formulation, with circle drawn. The unpaired nucleotides on the junction (blue circles) perfectly fit the circle by construction, while the paired nucleotides (green circles) do not. The purple line indicates the angle of the nucleotide 5' to the enclosing pair, whose optimal value is roughly 58°. (B) The line connecting two paired nucleotides is shown in purple, and different intersection points are shown, from 5' nucleotide (i = 0) to 3' (i = 1). The optimal value is roughly i = 0.3. (C) Two purple lines mark the distance _d_n from the circle's center to a base-paired nucleotide, and _d_a from the circle's center to the adjacent nucleotide. Each of the four base-paired nucleotides in this example will contribute a term (_d_n-_d_a)2 to the objective function. (D) Illustration of second formulation, using an example chosen so that some nucleotides would deviate significantly from the main circle. This circle is again shown explicitly. (E) The lower right unpaired region deviates from the main circle, but is positioned along an independent circle, which is shown. The four purple lines indicate deviations from this independent circle to the target main circle. Each of these lines corresponds to a term in the objective function.
Similar articles
- RNAconTest: comparing tools for noncoding RNA multiple sequence alignment based on structural consistency.
Wright ES. Wright ES. RNA. 2020 May;26(5):531-540. doi: 10.1261/rna.073015.119. Epub 2020 Jan 31. RNA. 2020. PMID: 32005745 Free PMC article. - ncRNA consensus secondary structure derivation using grammar strings.
Achawanantakun R, Sun Y, Takyar SS. Achawanantakun R, et al. J Bioinform Comput Biol. 2011 Apr;9(2):317-37. doi: 10.1142/s0219720011005501. J Bioinform Comput Biol. 2011. PMID: 21523935 - RNA2Drawer: geometrically strict drawing of nucleic acid structures with graphical structure editing and highlighting of complementary subsequences.
Johnson PZ, Kasprzak WK, Shapiro BA, Simon AE. Johnson PZ, et al. RNA Biol. 2019 Dec;16(12):1667-1671. doi: 10.1080/15476286.2019.1659081. Epub 2019 Aug 26. RNA Biol. 2019. PMID: 31441369 Free PMC article. - Sequence and structure analysis of noncoding RNAs.
Washietl S. Washietl S. Methods Mol Biol. 2010;609:285-306. doi: 10.1007/978-1-60327-241-4_17. Methods Mol Biol. 2010. PMID: 20221926 Review. - Energy-based RNA consensus secondary structure prediction in multiple sequence alignments.
Washietl S, Bernhart SH, Kellis M. Washietl S, et al. Methods Mol Biol. 2014;1097:125-41. doi: 10.1007/978-1-62703-709-9_7. Methods Mol Biol. 2014. PMID: 24639158 Review.
Cited by
- Bridge RNAs direct programmable recombination of target and donor DNA.
Durrant MG, Perry NT, Pai JJ, Jangid AR, Athukoralage JS, Hiraizumi M, McSpedon JP, Pawluk A, Nishimasu H, Konermann S, Hsu PD. Durrant MG, et al. Nature. 2024 Jun;630(8018):984-993. doi: 10.1038/s41586-024-07552-4. Epub 2024 Jun 26. Nature. 2024. PMID: 38926615 Free PMC article. - A rare bacterial RNA motif is implicated in the regulation of the purF gene whose encoded enzyme synthesizes phosphoribosylamine.
Malkowski SN, Atilho RM, Greenlee EB, Weinberg CE, Breaker RR. Malkowski SN, et al. RNA. 2020 Dec;26(12):1838-1846. doi: 10.1261/rna.077313.120. Epub 2020 Aug 25. RNA. 2020. PMID: 32843366 Free PMC article. - HOTAIR forms an intricate and modular secondary structure.
Somarowthu S, Legiewicz M, Chillón I, Marcia M, Liu F, Pyle AM. Somarowthu S, et al. Mol Cell. 2015 Apr 16;58(2):353-61. doi: 10.1016/j.molcel.2015.03.006. Epub 2015 Apr 9. Mol Cell. 2015. PMID: 25866246 Free PMC article. - Evaluating our ability to predict the structural disruption of RNA by SNPs.
Ritz J, Martin JS, Laederach A. Ritz J, et al. BMC Genomics. 2012 Jun 18;13 Suppl 4(Suppl 4):S6. doi: 10.1186/1471-2164-13-S4-S6. BMC Genomics. 2012. PMID: 22759654 Free PMC article. - A Branched SELEX Approach Identifies RNA Aptamers That Bind Distinct HIV-1 Capsid Structural Components.
Gruenke PR, Mayer MD, Aneja R, Schulze WJ, Song Z, Burke DH, Heng X, Lange MJ. Gruenke PR, et al. ACS Infect Dis. 2024 Aug 9;10(8):2637-2655. doi: 10.1021/acsinfecdis.3c00708. Epub 2024 Jul 17. ACS Infect Dis. 2024. PMID: 39016538 Free PMC article.
References
- Hüttenhofer A, Schattner P, Polacek N. Non-coding RNAs: hope or hype? Trends Genet. 2005;21:289–297. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources