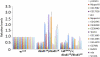Functional antagonism between histone H3K4 demethylases in vivo - PubMed (original) (raw)
Functional antagonism between histone H3K4 demethylases in vivo
Luisa Di Stefano et al. Genes Dev. 2011.
Abstract
Dynamic regulation of histone modifications is critical during development, and aberrant activity of chromatin-modifying enzymes has been associated with diseases such as cancer. Histone demethylases have been shown to play a key role in eukaryotic gene transcription; however, little is known about how their activities are coordinated in vivo to regulate specific biological processes. In Drosophila, two enzymes, dLsd1 (Drosophila ortholog of lysine-specific demethylase 1) and Lid (little imaginal discs), demethylate histone H3 at Lys 4 (H3K4), a residue whose methylation is associated with actively transcribed genes. Our studies show that compound mutation of Lid and dLsd1 results in increased H3K4 methylation levels. However, unexpectedly, Lid mutations strongly suppress dLsd1 mutant phenotypes. Investigation of the basis for this antagonism revealed that Lid opposes the functions of dLsd1 and the histone methyltransferase Su(var)3-9 in promoting heterochromatin spreading at heterochromatin-euchromatin boundaries. Moreover, our data reveal a novel role for dLsd1 in Notch signaling in Drosophila, and a complex network of interactions between dLsd1, Lid, and Notch signaling at euchromatic genes. These findings illustrate the complexity of functional interplay between histone demethylases in vivo, providing insights into the epigenetic regulation of heterochromatin/euchromatin boundaries by Lid and dLsd1 and showing their involvement in Notch pathway-specific control of gene expression in euchromatin.
Figures
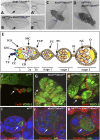
Figure 1.
Lid10424 mutation suppresses phenotypes associated with dLsd1ΔN loss-of-function mutation. (A) Loss of dLsd1 results in extra wing vein tissue emanating from the pcv (indicated by arrows; closer view in A′). (B) This phenotype is rescued by heterozygous mutation of Lid (indicated by arrows; closer view in B′). (C,D) Lid mutation suppresses the held-out wing phenotype observed in homozygous dLsd1 mutant flies. (E) Schematic representation of a wild-type ovariole, including the germarium and egg chambers (stages 1–3). Germline stem cells (GSC) reside at the tip of the germarium in a microenvironment created by the cap cells (CC) and the terminal filaments (TF). The differentiating daughter cell of a germline stem cell is the cystoblast (CB), which moves posterior and becomes encompassed by inner germarian sheath cells (IGS). Cystoblasts divide four times and generate germline cysts of 16 cells, which, after passing region 2 of the germarium, become surrounded by follicle cell precursors (FCP). Follicle cell precursors are generated by somatic stem cells (SSC) and differentiate into follicle cells (FC), polar cells (PC), and stalk cells (SC). Among the 16 germ cells, one differentiates into the oocyte (O), and the remaining 15 become nurse cells (NC). (F,G,H) Lid10424 mutations partially rescue dLsd1ΔN mutant follicle cell defects. Wild-type (w1118), dLsd1ΔN homozygous, and Lid10424/+; dLsd1ΔN/dLsd1ΔN ovaries were stained with YOYO-1 (green) and anti-Fasciclin-III to outline somatic cells. (I,J,K) Lid10424 mutation partially rescues dLsd1ΔN mutant defects in the germline. Wild-type (w1118), dLsd1ΔN homozygous, and Lid10424/+; dLsd1ΔN/dLsd1ΔN ovaries were stained with TOTO (blue) and anti-orb to visualize the oocyte (indicated by arrows). Cell outlines and ring canals were visualized by phalloidin staining of the actin cytoskeleton (red).
Figure 2.
dLsd1 and Lid mutations cooperatively increase the global level of histone H3K4 methylation. (A) Increase in histone H3K4 methylation level in double-mutant adult flies. Immunoblots of wild-type (wt) versus HDM mutant lysates from adult flies were probed with antibodies specific for mono-, di-, and trimethyl H3K4 and pan-acetyl H3; anti-H3 was used as a loading control. (B–G) H3K4 methylation levels are increased in polytene chromosomes of double mutants. Staining of polytene chromosome with an antibody specific for H3K4me2 (red) and with YOYO-1 (green). Reduced levels of Lid in combination with loss of dLsd1 results in a chromosome-wide increase in H3K4me2 levels.
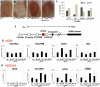
Figure 3.
Lid and dLsd1 antagonistically control heterochromatin spreading in PEV. (A–D) The effect of a reduced dosage of Lid and dLsd1 on wm4h variegation compared with control flies is shown. Reducing the dosage of dLsd1 results in suppression of variegation (B), while reducing the levels of Lid results in enhancement (C); combined reduction of Lid and dLsd1 dosage has a minimal effect on variegation (D). (E) The effect of a reduced dosage of Lid and dLsd1 on T(2;3)Sbv variegation compared with control flies is shown. The effect was quantified by counting the number of stubble and wild-type bristles of the thorax. A minimum of 380 bristles per genotype was counted. (F) A schematic representation of the white locus at the wm4 inversion is shown. (G) dLsd1 and Lid mutations have opposite effects on white gene expression. Reducing the levels of dLsd1 results in increased expression of the white gene, while reducing the levels of Lid results in decreased expression as compared with wild-type flies. Combined reduction of Lid and dLsd1 dosage results in an intermediate effect compared with single mutants. (H) dLsd1 and Lid control H3K9 methylation levels at the white locus. ChIP analysis of H3K9me2 levels along the wm4h inverted region. Reducing the levels of dLsd1 results in decreased levels of H3K9me2 at the white locus, while reducing the levels of Lid results in increased H3K9me2, and a combined reduction of Lid and dLsd1 dosage results in levels of H3K9me2 comparable with wild-type flies.
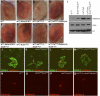
Figure 4.
Enhancement of variegation due to Lid mutation is dependent on Su(var)3–9 dosage, and Lid mutation results in increased levels of H3K9 methylation. (A–H) The enhancer effect of Lid mutation on white variegation in wm4 is suppressed by reduction of Su(var)3–9 dosage. (I) H3K9 methylation levels are globally increased in Lid mutants. Immunoblot analysis of global levels of H3K9me2 in adult males of the indicated genotypes. Total histone H3 and acetyl-H3 levels are shown. (J–Q) H3K9 methylation levels are increased at the chromocenter of polytene chromosomes in mutants for the Lid allele. The top panel shows the staining of polytene chromosomes with an antibody specific for H3K9me2 (red) and with YOYO-1 (green), whereas the bottom panel shows anti-H3K9me2 staining alone. dLsd1 mutation results in reduced levels of H3K9me2 at the chromocenter, while Lid mutation results in an increase of H3K9me2 at the chromocenter compared with w1118. Reduced levels of Lid in combination with loss of dLsd1 results in H3K9me2 levels comparable with wild-type flies.
Figure 5.
Increased expression of genes at the 2R euchromatin–heterochromatin boundary in dLsd1 mutant flies. RT-qPCR analysis of the expression of the indicated genes in w1118 (wild-type), dLsd1 mutant, and Lid and dLsd1 double mutant ovaries is shown. The expression level is normalized against the w1118, wild-type control. Experiments were performed in triplicate, and error bars indicate standard error of the mean. Supplemental Table S4 displays the sequence coordinates and the position on the cytogenetic map of each of the genes used in this expression analysis.
Figure 6.
dLsd1 directly regulates Notch target gene expression by controlling histone methylation levels. (A) dLsd1 is required for repression of Notch target genes. RT-qPCR analysis of the expression of E(spl) genes in w1118 (wild-type, wt) and dLsd1 mutant flies. The expression level is normalized against the w1118, wild-type control. Experiments were performed in triplicate, and error bars indicate the standard error of the mean. (B) dLsd1 binds directly to the E(spl) locus. ChIP analysis of dLsd1 binding across the E(spl) locus using an anti-dLsd1-specific antibody in S2 cells incubated with dsRNA against Luciferase (mock) and dLsd1 to control for specificity. Preimmune serum was used as an additional control for specificity. ChIP data are the result of three independent immunoprecipitations. Error bars indicate standard deviation. Each gene is represented by two or three fragments: enhancer (e), distal (d), and proximal (p) to the transcriptional start site. (C) dLsd1 and Lid cooperatively control histone methylation levels at Notch target genes. Cross-linked chromatin was isolated from S2 cells incubated with dsRNA against the indicated mRNAs, and ChIP analysis was performed using antibodies specific for H3K4me1, H3K4me2, and H3K4me3. Error bars indicate standard deviation.
Figure 7.
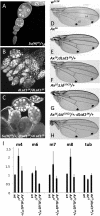
dLsd1 genetically interacts with various components of the Notch signaling pathway. (A–C) Su(H)05 partially suppresses the oogenesis defect in dLsd1 homozygous mutant flies. (D–H) dLsd1 mutation suppresses truncations of the L4 and L5 wing veins in flies heterozygous for the Ax16 gain-of-function Notch mutation, while Lid mutation enhances it. Shown are representative examples of wings of the indicated genotypes; arrows mark the distal part of L4 and L5 wing veins where the phenotype is evident. (I) dLsd1 is required for activation of Notch target genes. RT-qPCR analysis of the expression of E(spl) genes in w1118 (wild-type, wt), AxE2, and AxE2, dLsd1 heterozygous mutant males. The expression level is normalized against the w1118, wild-type control. Experiments were performed in triplicate, and error bars indicate standard error of the mean.
Similar articles
- Characterization of Drosophila melanogaster JmjC+N histone demethylases.
Lloret-Llinares M, Carré C, Vaquero A, de Olano N, Azorín F. Lloret-Llinares M, et al. Nucleic Acids Res. 2008 May;36(9):2852-63. doi: 10.1093/nar/gkn098. Epub 2008 Mar 29. Nucleic Acids Res. 2008. PMID: 18375980 Free PMC article. - Mutation of Drosophila Lsd1 disrupts H3-K4 methylation, resulting in tissue-specific defects during development.
Di Stefano L, Ji JY, Moon NS, Herr A, Dyson N. Di Stefano L, et al. Curr Biol. 2007 May 1;17(9):808-12. doi: 10.1016/j.cub.2007.03.068. Curr Biol. 2007. PMID: 17462898 Free PMC article. - Heterochromatin formation in Drosophila is initiated through active removal of H3K4 methylation by the LSD1 homolog SU(VAR)3-3.
Rudolph T, Yonezawa M, Lein S, Heidrich K, Kubicek S, Schäfer C, Phalke S, Walther M, Schmidt A, Jenuwein T, Reuter G. Rudolph T, et al. Mol Cell. 2007 Apr 13;26(1):103-15. doi: 10.1016/j.molcel.2007.02.025. Mol Cell. 2007. PMID: 17434130 - The function and regulation of the JARID1 family of histone H3 lysine 4 demethylases: the Myc connection.
Secombe J, Eisenman RN. Secombe J, et al. Cell Cycle. 2007 Jun 1;6(11):1324-8. doi: 10.4161/cc.6.11.4269. Epub 2007 Jun 14. Cell Cycle. 2007. PMID: 17568193 Review. - Current perspectives on histone demethylases.
Tian X, Fang J. Tian X, et al. Acta Biochim Biophys Sin (Shanghai). 2007 Feb;39(2):81-8. doi: 10.1111/j.1745-7270.2007.00272.x. Acta Biochim Biophys Sin (Shanghai). 2007. PMID: 17277881 Review.
Cited by
- Physical and functional interactions between the histone H3K4 demethylase KDM5A and the nucleosome remodeling and deacetylase (NuRD) complex.
Nishibuchi G, Shibata Y, Hayakawa T, Hayakawa N, Ohtani Y, Sinmyozu K, Tagami H, Nakayama J. Nishibuchi G, et al. J Biol Chem. 2014 Oct 17;289(42):28956-70. doi: 10.1074/jbc.M114.573725. Epub 2014 Sep 4. J Biol Chem. 2014. PMID: 25190814 Free PMC article. - Notch-dependent and -independent functions of transcription factor RBPJ.
Friedrich T, Ferrante F, Pioger L, Nist A, Stiewe T, Andrau JC, Bartkuhn M, Giaimo BD, Borggrefe T. Friedrich T, et al. Nucleic Acids Res. 2022 Aug 12;50(14):7925-7937. doi: 10.1093/nar/gkac601. Nucleic Acids Res. 2022. PMID: 35848919 Free PMC article. - Jarid1b targets genes regulating development and is involved in neural differentiation.
Schmitz SU, Albert M, Malatesta M, Morey L, Johansen JV, Bak M, Tommerup N, Abarrategui I, Helin K. Schmitz SU, et al. EMBO J. 2011 Nov 16;30(22):4586-600. doi: 10.1038/emboj.2011.383. EMBO J. 2011. PMID: 22020125 Free PMC article. - Drosophila Set1 is the major histone H3 lysine 4 trimethyltransferase with role in transcription.
Ardehali MB, Mei A, Zobeck KL, Caron M, Lis JT, Kusch T. Ardehali MB, et al. EMBO J. 2011 Jun 21;30(14):2817-28. doi: 10.1038/emboj.2011.194. EMBO J. 2011. PMID: 21694722 Free PMC article. - Functions and Interactions of Mammalian KDM5 Demethylases.
Pavlenko E, Ruengeler T, Engel P, Poepsel S. Pavlenko E, et al. Front Genet. 2022 Jul 11;13:906662. doi: 10.3389/fgene.2022.906662. eCollection 2022. Front Genet. 2022. PMID: 35899196 Free PMC article. Review.
References
- Bannister AJ, Kouzarides T 2005. Reversing histone methylation. Nature 436: 1103–1106 - PubMed
- Bernstein BE, Kamal M, Lindblad-Toh K, Bekiranov S, Bailey DK, Huebert DJ, McMahon S, Karlsson EK, Kulbokas EJ 3rd, Gingeras TR, et al. 2005. Genomic maps and comparative analysis of histone modifications in human and mouse. Cell 120: 169–181 - PubMed
- Bray SJ 2006. Notch signalling: A simple pathway becomes complex. Nat Rev Mol Cell Biol 7: 678–689 - PubMed
- Burgio G, La Rocca G, Sala A, Arancio W, Di Gesù D, Collesano M, Sperling AS, Armstrong JA, van Heeringen SJ, Logie C, et al. 2008. Genetic identification of a network of factors that functionally interact with the nucleosome remodeling ATPase ISWI. PLoS Genet 4: e1000089 doi: 10.1371/journal.pgen.1000089 - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 GM071449/GM/NIGMS NIH HHS/United States
- R01 GM053203/GM/NIGMS NIH HHS/United States
- GM81607/GM/NIGMS NIH HHS/United States
- GM071449/GM/NIGMS NIH HHS/United States
- CA64402/CA/NCI NIH HHS/United States
- R01 CA064402/CA/NCI NIH HHS/United States
- GM53203/GM/NIGMS NIH HHS/United States
- TCR09002/TI_/Telethon/Italy
- R01 GM081607/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous