Galaxy CloudMan: delivering cloud compute clusters - PubMed (original) (raw)
Galaxy CloudMan: delivering cloud compute clusters
Enis Afgan et al. BMC Bioinformatics. 2010.
Abstract
Background: Widespread adoption of high-throughput sequencing has greatly increased the scale and sophistication of computational infrastructure needed to perform genomic research. An alternative to building and maintaining local infrastructure is "cloud computing", which, in principle, offers on demand access to flexible computational infrastructure. However, cloud computing resources are not yet suitable for immediate "as is" use by experimental biologists.
Results: We present a cloud resource management system that makes it possible for individual researchers to compose and control an arbitrarily sized compute cluster on Amazon's EC2 cloud infrastructure without any informatics requirements. Within this system, an entire suite of biological tools packaged by the NERC Bio-Linux team (http://nebc.nerc.ac.uk/tools/bio-linux) is available for immediate consumption. The provided solution makes it possible, using only a web browser, to create a completely configured compute cluster ready to perform analysis in less than five minutes. Moreover, we provide an automated method for building custom deployments of cloud resources. This approach promotes reproducibility of results and, if desired, allows individuals and labs to add or customize an otherwise available cloud system to better meet their needs.
Conclusions: The expected knowledge and associated effort with deploying a compute cluster in the Amazon EC2 cloud is not trivial. The solution presented in this paper eliminates these barriers, making it possible for researchers to deploy exactly the amount of computing power they need, combined with a wealth of existing analysis software, to handle the ongoing data deluge.
Figures
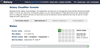
Figure 1
Main web interface for Galaxy CloudMan. Screenshot of the CloudMan cloud controller web interface running on the master instance of the cloud compute cluster. This interface is used to control the size of the cloud cluster, including adding cluster services, scaling the size of the cluster in terms of worker instances and associated persistent data volume, and as an overview of the cluster status.
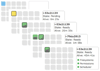
Figure 2
Scaling worker instances within CloudMan. A progression of the act of scaling the number of worker instances associated with the given cloud cluster. Each icon represents an individual cloud instance. Within each icon, the load of the instance over the past 15 minutes is shown as a small glyph. Based on the load of worker instances, the user can decide to scale the size of the cluster up or down.
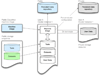
Figure 3
Modular architecture of CloudMan. The architecture of CloudMan is based on separation and subsequent coordination of otherwise independent components: the machine image, a persistent data repository, and persistent storage resources (i.e., snapshots). The machine image is characterized by simplicity; it consists only of the basic services required to initiate the application unit deployment process. The persistent data repository lives independent of the machine image and is used to provide instance contextualization details, such as, boot time scripts that define which services should be started. Lastly, persistent storage resources or snapshots are used as the storage medium for tools, libraries, or datasets required by the tools. Once instantiated, those components are aggregated by CloudMan into a cohesive operational unit. Because they are not modified during the life of a cluster, the persistent storage resources are deleted upon cluster termination. All of the cluster settings and user data are preserved in user’s account and will be reused on next cluster instantiation. The Galaxy CloudMan developers maintain items in blue; items in green are (currently) maintained by the CloudBioLinux community (
), while items in gray are private to a user.
Similar articles
- Using cloud computing infrastructure with CloudBioLinux, CloudMan, and Galaxy.
Afgan E, Chapman B, Jadan M, Franke V, Taylor J. Afgan E, et al. Curr Protoc Bioinformatics. 2012 Jun;Chapter 11:11.9.1-11.9.20. doi: 10.1002/0471250953.bi1109s38. Curr Protoc Bioinformatics. 2012. PMID: 22700313 Free PMC article. - Genomics Virtual Laboratory: A Practical Bioinformatics Workbench for the Cloud.
Afgan E, Sloggett C, Goonasekera N, Makunin I, Benson D, Crowe M, Gladman S, Kowsar Y, Pheasant M, Horst R, Lonie A. Afgan E, et al. PLoS One. 2015 Oct 26;10(10):e0140829. doi: 10.1371/journal.pone.0140829. eCollection 2015. PLoS One. 2015. PMID: 26501966 Free PMC article. - Laniakea: an open solution to provide Galaxy "on-demand" instances over heterogeneous cloud infrastructures.
Tangaro MA, Donvito G, Antonacci M, Chiara M, Mandreoli P, Pesole G, Zambelli F. Tangaro MA, et al. Gigascience. 2020 Apr 1;9(4):giaa033. doi: 10.1093/gigascience/giaa033. Gigascience. 2020. PMID: 32252069 Free PMC article. - User-centric genomics infrastructure: trends and technologies.
Krishna R, Elisseev V. Krishna R, et al. Genome. 2021 Apr;64(4):467-475. doi: 10.1139/gen-2020-0096. Epub 2020 Nov 20. Genome. 2021. PMID: 33216660 Review. - Bioinformatics clouds for big data manipulation.
Dai L, Gao X, Guo Y, Xiao J, Zhang Z. Dai L, et al. Biol Direct. 2012 Nov 28;7:43; discussion 43. doi: 10.1186/1745-6150-7-43. Biol Direct. 2012. PMID: 23190475 Free PMC article. Review.
Cited by
- Advances in Faba Bean Genetics and Genomics.
O'Sullivan DM, Angra D. O'Sullivan DM, et al. Front Genet. 2016 Aug 22;7:150. doi: 10.3389/fgene.2016.00150. eCollection 2016. Front Genet. 2016. PMID: 27597858 Free PMC article. Review. - Using cloud computing infrastructure with CloudBioLinux, CloudMan, and Galaxy.
Afgan E, Chapman B, Jadan M, Franke V, Taylor J. Afgan E, et al. Curr Protoc Bioinformatics. 2012 Jun;Chapter 11:11.9.1-11.9.20. doi: 10.1002/0471250953.bi1109s38. Curr Protoc Bioinformatics. 2012. PMID: 22700313 Free PMC article. - Making whole genome multiple alignments usable for biologists.
Blankenberg D, Taylor J, Nekrutenko A; Galaxy Team. Blankenberg D, et al. Bioinformatics. 2011 Sep 1;27(17):2426-8. doi: 10.1093/bioinformatics/btr398. Epub 2011 Jul 19. Bioinformatics. 2011. PMID: 21775304 Free PMC article. - Experiences with workflows for automating data-intensive bioinformatics.
Spjuth O, Bongcam-Rudloff E, Hernández GC, Forer L, Giovacchini M, Guimera RV, Kallio A, Korpelainen E, Kańduła MM, Krachunov M, Kreil DP, Kulev O, Łabaj PP, Lampa S, Pireddu L, Schönherr S, Siretskiy A, Vassilev D. Spjuth O, et al. Biol Direct. 2015 Aug 19;10:43. doi: 10.1186/s13062-015-0071-8. Biol Direct. 2015. PMID: 26282399 Free PMC article. - Genomics Virtual Laboratory: A Practical Bioinformatics Workbench for the Cloud.
Afgan E, Sloggett C, Goonasekera N, Makunin I, Benson D, Crowe M, Gladman S, Kowsar Y, Pheasant M, Horst R, Lonie A. Afgan E, et al. PLoS One. 2015 Oct 26;10(10):e0140829. doi: 10.1371/journal.pone.0140829. eCollection 2015. PLoS One. 2015. PMID: 26501966 Free PMC article.
References
- Armbrust M, Fox A, Griffith R, Joseph AD, Katz R, Konwinski A, Lee G, Patterson D, Rabkin A, Stoica I, Zaharia M. In: Book Above the Clouds: A Berkeley View of Cloud Computing. Editor ed.^eds., editor. University of California at Berkeley; 2009. Above the Clouds: A Berkeley View of Cloud Computing; p. 23.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources