Flexible taxonomic assignment of ambiguous sequencing reads - PubMed (original) (raw)
Flexible taxonomic assignment of ambiguous sequencing reads
José C Clemente et al. BMC Bioinformatics. 2011.
Abstract
Background: To characterize the diversity of bacterial populations in metagenomic studies, sequencing reads need to be accurately assigned to taxonomic units in a given reference taxonomy. Reads that cannot be reliably assigned to a unique leaf in the taxonomy (ambiguous reads) are typically assigned to the lowest common ancestor of the set of species that match it. This introduces a potentially severe error in the estimation of bacteria present in the sample due to false positives, since all species in the subtree rooted at the ancestor are implicitly assigned to the read even though many of them may not match it.
Results: We present a method that maps each read to a node in the taxonomy that minimizes a penalty score while balancing the relevance of precision and recall in the assignment through a parameter q. This mapping can be obtained in time linear in the number of matching sequences, because LCA queries to the reference taxonomy take constant time. When applied to six different metagenomic datasets, our algorithm produces different taxonomic distributions depending on whether coverage or precision is maximized. Including information on the quality of the reads reduces the number of unassigned reads but increases the number of ambiguous reads, stressing the relevance of our method. Finally, two measures of performance are described and results with a set of artificially generated datasets are discussed.
Conclusions: The assignment strategy of sequencing reads introduced in this paper is a versatile and a quick method to study bacterial communities. The bacterial composition of the analyzed samples can vary significantly depending on how ambiguous reads are assigned depending on the value of the q parameter. Validation of our results in an artificial dataset confirm that a combination of values of q produces the most accurate results.
Figures
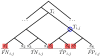
Figure 1
Sample Taxonomic Assignment of an Ambiguous Read. Assigning read Ri to the _j_th node of Ti partitions the leaves of Ti into true positives, false positives, true negatives, and false negatives. In this example, the hits are Mi = {_s_1, _s_5, _s_6, _s_7}. If we let j be the blue circled node, we obtain TPi,j = {_s_5, _s_6, _s_7}, FPi,j = {_s_8}, TNi,j = {_s_2, _s_3, _s_4}, FNi,j = {_s_1}. True hit H = _s_5 (defined in Section "Validation: Performance in ROC Space").
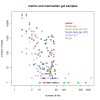
Figure 2
Validation of Results in ROC Space. Distance in ROC space to the diagonal TPRH = FPRH. Points above the diagonal represent good predictions, the larger the distance the better.
Figure 3
Distribution of Sequencing Reads. Distribution of sequencing reads (number of hits and reads) ambiguously assigned with up to 2 mismatches to two or more of the 5,165 sequences in the reference bacterial taxonomy.
Figure 4
Distribution of Hits per Taxonomic Rank. Distribution of the number of hits (species with up to k mismatches) in ambiguous reads per taxonomic rank.
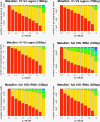
Figure 5
Distribution of Taxonomic Ranks in Metagenomic Datasets. Ambiguous reads assigned in the bacterial taxonomy at each taxonomic rank for q = 0, ..., 1. Color code: domain: purple; phylum: indigo; class: light blue; order: cyan; family: green; genus: yellow; species: red.
Figure 6
Distribution of Taxonomic Ranks in Simulated Datasets. Simulated ambiguous reads assigned in the bacterial taxonomy at each taxonomic rank for q = 0, ..., 1. Datasets were constructed from whole 16S rRNA sequence and from the V1-V2 hypervariable region. Color code: domain: purple; phylum: indigo; class: light blue; order: cyan; family: green; genus: yellow; species: red.
Similar articles
- Accurate taxonomic assignment of short pyrosequencing reads.
Clemente JC, Jansson J, Valiente G. Clemente JC, et al. Pac Symp Biocomput. 2010:3-9. doi: 10.1142/9789814295291_0002. Pac Symp Biocomput. 2010. PMID: 19908352 - MinION™ nanopore sequencing of environmental metagenomes: a synthetic approach.
Brown BL, Watson M, Minot SS, Rivera MC, Franklin RB. Brown BL, et al. Gigascience. 2017 Mar 1;6(3):1-10. doi: 10.1093/gigascience/gix007. Gigascience. 2017. PMID: 28327976 Free PMC article. - Pseudoalignment for metagenomic read assignment.
Schaeffer L, Pimentel H, Bray N, Melsted P, Pachter L. Schaeffer L, et al. Bioinformatics. 2017 Jul 15;33(14):2082-2088. doi: 10.1093/bioinformatics/btx106. Bioinformatics. 2017. PMID: 28334086 Free PMC article. - Sequence assembly using next generation sequencing data--challenges and solutions.
Chin FY, Leung HC, Yiu SM. Chin FY, et al. Sci China Life Sci. 2014 Nov;57(11):1140-8. doi: 10.1007/s11427-014-4752-9. Epub 2014 Oct 17. Sci China Life Sci. 2014. PMID: 25326069 Review. - Assessment of metagenomic assemblers based on hybrid reads of real and simulated metagenomic sequences.
Wang Z, Wang Y, Fuhrman JA, Sun F, Zhu S. Wang Z, et al. Brief Bioinform. 2020 May 21;21(3):777-790. doi: 10.1093/bib/bbz025. Brief Bioinform. 2020. PMID: 30860572 Free PMC article. Review.
Cited by
- Phylogenetic placement of metagenomic reads using the minimum evolution principle.
Filipski A, Tamura K, Billing-Ross P, Murillo O, Kumar S. Filipski A, et al. BMC Genomics. 2015;16 Suppl 1(Suppl 1):S13. doi: 10.1186/1471-2164-16-S1-S13. Epub 2015 Jan 15. BMC Genomics. 2015. PMID: 25923672 Free PMC article. - BioMaS: a modular pipeline for Bioinformatic analysis of Metagenomic AmpliconS.
Fosso B, Santamaria M, Marzano M, Alonso-Alemany D, Valiente G, Donvito G, Monaco A, Notarangelo P, Pesole G. Fosso B, et al. BMC Bioinformatics. 2015 Jul 1;16:203. doi: 10.1186/s12859-015-0595-z. BMC Bioinformatics. 2015. PMID: 26130132 Free PMC article. - Analytical tools and databases for metagenomics in the next-generation sequencing era.
Kim M, Lee KH, Yoon SW, Kim BS, Chun J, Yi H. Kim M, et al. Genomics Inform. 2013 Sep;11(3):102-13. doi: 10.5808/GI.2013.11.3.102. Epub 2013 Sep 30. Genomics Inform. 2013. PMID: 24124405 Free PMC article. Review. - Statistical approach of functional profiling for a microbial community.
An L, Pookhao N, Jiang H, Xu J. An L, et al. PLoS One. 2014 Sep 8;9(9):e106588. doi: 10.1371/journal.pone.0106588. eCollection 2014. PLoS One. 2014. PMID: 25198674 Free PMC article. - Metagenomic Classification Using an Abstraction Augmented Markov Model.
Zhu XS, McGee M. Zhu XS, et al. J Comput Biol. 2016 Feb;23(2):111-122. doi: 10.1089/cmb.2015.0141. Epub 2015 Nov 30. J Comput Biol. 2016. PMID: 26618474 Free PMC article.
References
- Alberts B, Johnson A, Lewis J, Raff M, Roberts K, Walter P. Molecular Biology of the Cell. 5. New York, USA: Garland Science; 2008.
- Gray NF. Biology of Wastewater Treatment. 2. London, UK: Imperial College Press; 2004.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials