Massive genomic rearrangement acquired in a single catastrophic event during cancer development - PubMed (original) (raw)
. 2011 Jan 7;144(1):27-40.
doi: 10.1016/j.cell.2010.11.055.
Chris D Greenman, Beiyuan Fu, Fengtang Yang, Graham R Bignell, Laura J Mudie, Erin D Pleasance, King Wai Lau, David Beare, Lucy A Stebbings, Stuart McLaren, Meng-Lay Lin, David J McBride, Ignacio Varela, Serena Nik-Zainal, Catherine Leroy, Mingming Jia, Andrew Menzies, Adam P Butler, Jon W Teague, Michael A Quail, John Burton, Harold Swerdlow, Nigel P Carter, Laura A Morsberger, Christine Iacobuzio-Donahue, George A Follows, Anthony R Green, Adrienne M Flanagan, Michael R Stratton, P Andrew Futreal, Peter J Campbell
Affiliations
- PMID: 21215367
- PMCID: PMC3065307
- DOI: 10.1016/j.cell.2010.11.055
Massive genomic rearrangement acquired in a single catastrophic event during cancer development
Philip J Stephens et al. Cell. 2011.
Abstract
Cancer is driven by somatically acquired point mutations and chromosomal rearrangements, conventionally thought to accumulate gradually over time. Using next-generation sequencing, we characterize a phenomenon, which we term chromothripsis, whereby tens to hundreds of genomic rearrangements occur in a one-off cellular crisis. Rearrangements involving one or a few chromosomes crisscross back and forth across involved regions, generating frequent oscillations between two copy number states. These genomic hallmarks are highly improbable if rearrangements accumulate over time and instead imply that nearly all occur during a single cellular catastrophe. The stamp of chromothripsis can be seen in at least 2%-3% of all cancers, across many subtypes, and is present in ∼25% of bone cancers. We find that one, or indeed more than one, cancer-causing lesion can emerge out of the genomic crisis. This phenomenon has important implications for the origins of genomic remodeling and temporal emergence of cancer.
Copyright © 2011 Elsevier Inc. All rights reserved.
Figures
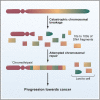
Graphical abstract
Figure 1
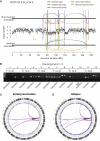
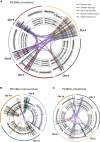
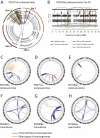
Clustered Rearrangements on Chromosome 4q in a Patient with Chronic Lymphocytic Leukemia (A) Copy number between 70 Mb and 170 Mb of the chromosome oscillates between a copy number of 1 and 2, demarcated by back-and-forth intrachromosomal rearrangements of all four possible orientations, as well as several interchromosomal rearrangements. (B) PCR gel of 12 putative genomic rearrangements identified by sequencing. PCR across the breakpoint is performed for each rearrangement on tumor DNA for samples taken at initial presentation (T1) and relapse (T2) as well as germline DNA (N). (C) Genome-wide profile of rearrangements in a sample taken before chemotherapy. Chromosomes range round the outside of the circle, copy number changes are shown by the blue line in the inner ring, and somatically acquired genomic rearrangements are shown as arcs linking the two relevant genomic points. (D) Genome-wide profile of rearrangements from the same patient 31 months later, at relapse after therapy.
Figure 2
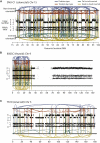
Rearrangement Screens in Three Cancer Cell Lines Showing Evidence for Chromothripsis Copy number profiles derive from SNP6 microarray data and are shown as the upper panel of points for each cell line. Allelic ratios for each SNP are shown in the lower panel of dots: homozygous SNPs cluster at allelic ratios near 0 or 1, heterozygous SNPs cluster around 0.5. Intrachromosomal rearrangements of all four possible orientations are shown, with deletion-type events as blue lines, tandem duplication-type in red, tail-to-tail inverted rearrangements in green and head-to-head inverted rearrangements in yellow. (A) SNU-C1, a cell line from a colorectal cancer, carries 239 rearrangements involving chromosome 15. (B) 8505C, a thyroid cancer cell line, has 77 rearrangements involving chromosome 9p. (C) TK10, a renal cancer cell line, has 55 rearrangements involving chromosome 5.
Figure 3
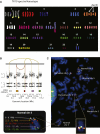
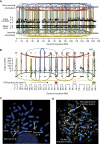
FISH Profiling of TK10 (A) The spectral karyotype of the TK10 genome. (B) Copy number profiles and genomic rearrangements for the five regions of chromosome 5 studied by multicolor FISH. (C) Multicolor FISH of TK10.
Figure 4
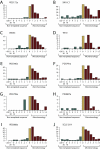
Chromothripsis Involving More Than One Chromosome in Primary Samples from Patients with Bone Cancer For each case, the relevant chromosomes are shown with SNP6 microarray copy number profiles in the outer ring, allelic ratios in the inner ring, and somatically acquired genomic rearrangements shown as arcs in the center. (A) PD3808a, from a chordoma, shows 147 rearrangements interlinking chromosomes 3q, 4q, 7q, 8p, and 9p. (B) PD3786a, an osteosarcoma sample, carries 88 rearrangements involving chromosome 8, 12, and 14. (C) PD3807a, another chordoma sample, has 38 rearrangements involving chromosomes 1p, 3, 8, and 14.
Figure 5
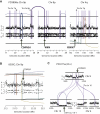
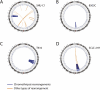
Genomic Features of Chromothripsis Suggest that Most Rearrangements Occur in a Single Catastrophic Event (A) Example of a sequence of progressive rearrangements disrupting a model chromosome. The chromosomal configuration after each rearrangement is shown, together with the copy number and rearrangement plot that would result (in the style of Figure 2). (B) Example of how a chromosomal catastrophe might break the chromosome into many pieces that are then stitched back together haphazardly. (C) One thousand Monte Carlo simulations (black points) performed under the assumption that rearrangements accumulate progressively over time show the number of copy number states seen in the resultant derivative chromosome. Samples with chromothripsis, shown as red diamonds, fall well outside this spectrum. (D) Observed distances between adjacent breakpoints for each sample are shown beside the expected distribution if breaks occurred in entirely random locations.
Figure 6
Generation of a Double Minute Chromosome Containing MYC by Chromothripsis in a Small Cell Lung Cancer Cell Line, SCLC-21H (A) Copy number profile, allelic ratio, and rearrangements of chromosome 8. (B) Copy number data from the rearrangement screen shows 15 discrete regions of chromosome 8 that are massively amplified, with 50–200 copies per cell. Each amplified region is demarcated by rearrangements linking to other heavily amplified segments (thick lines), with evidence for later internal rearrangements also found (thin lines). (C) Three color FISH for three regions of chromosome 8 (predicted to be linked by the rearrangement data, but not amplified; green, 13 Mb; red, 41 Mb; pale pink, 49 Mb). (D) FISH for three heavily amplified regions. The locations of the probes are shown in Figure 6B (red, 66.5 Mb; white, 99.3 Mb; green, 128.8 Mb).
Figure 7
Loss of Tumor Suppressor Genes through Chromothripsis (A) PD3808a, the chordoma sample shown in Figure 4A, shows clustered chromothripsis rearrangements around CDKN2A, leading to a loss of one copy of this tumor suppressor gene. The other copy is also lost, through a deletion, which presumably occurred on the other parental copy of chromosome 9p at a separate time-point (thick line). The same cluster of chromothripsis rearrangements causes loss of a second tumor suppressor gene, FBXW7, on chromosome 4q and a third cancer gene, WRN, on chromosome 8p. (B) Chromothripsis has also led to loss of one copy of CDKN2A in the thyroid cancer cell line, 8505C. (C) Loss of two tumor suppressor genes, CDKN2A and the microRNA cluster miR-15a/16-1, by clustered rearrangements involving chromosomes 4, 9, and 13 in a patient with CLL, PD3175a.
Figure S2
Genome-wide Rearrangements for Four Cell Lines with Chromothripsis, Related to Figure 2 (A–D) Circos plots for (A) SNU-C1, (B) 8505C, (C) TK10 and (D) SCLC-21H. Around the outside are ideograms of the chromosome, with the inner ring representing copy number segments. Chromosomal rearrangements are shown as arcs in the middle joining the two relevant regions of the genome for each rearrangement. Chromothripsis rearrangements are shown in blue and those not associated with chromothripsis in orange.
Figure S3
Spectral Karyotypes for (A) 8505C and (B) SNU-C1, Related to Figure 3
Figure S4
Chromothripsis Involving Several Chromosomes, Related to Figure 4 (A) PD3791a, with osteosarcoma, shows 86 rearrangements involving chromosomes 7, 9, 12 and 13, with SNP6 microarray copy number profiles in the outer ring, allelic ratios in the inner ring and somatically acquired genomic rearrangements shown as arcs in the centre. (B) PD3799a, also from a patient with osteosarcoma, shows 28 rearrangements involving chromosome 20. (C–H) Circos plots for (C) PD3786a, (D) PD3791a, (E) PD3799a, (F) PD3807a, (G) PD3808a and (H) PD3646a. Around the outside are ideograms of the chromosome, with the inner ring representing copy number segments. Chromosomal rearrangements are shown as arcs in the middle joining the two relevant regions of the genome for each rearrangement. Chromothripsis rearrangements are shown in blue and those not associated with chromothripsis in orange.
Figure S5
Signatures of DNA Repair at the Breakpoint, Related to Figure 5 Patterns of microhomology (red), non-templated sequence (teal) or direct end-joining (yellow) in the chromothripsis rearrangements for each sample. The x axis shows the number of bases of microhomology (right of 0) or non-templated sequence (left of 0) for each rearrangement. The y axis shows the number of rearrangements in the sample showing that pattern of microhomology or non-templated sequence.
Figure S6
Marker Chromosomes Created by Chromothripsis, Related to Figure 6 (A) Multicolor FISH for the three heavily amplified probes in SCLC-21H (shown in Figure 6B) together with whole chromosome paint for chromosome 8 (purple). The normal chromosome 8 has the three probes in the correct orientation, whereas the probes do not hybridize to the two derivative chromosomes. (B) A rearrangement screen from a pancreatic cancer, PD3646a, previously published (Campbell et al., 2010), shows 41 rearrangements involving chromosomes 1, 4, 10 and 14. Copy number profiles for the relevant chromosomes are in the outer ring with somatically acquired genomic rearrangements shown as arcs in the centre. (C) Spectral karyotype for PD3646a shows a striped marker chromosome, with at least 6 cytogenetically visible segments arising from multiple chromosomes. The bright yellow signal corresponds to paint from chromosome 22, but the other stripes are characteristic of signals from chromosomes 1, 4, 10 and 14.
Figure S7
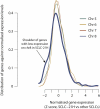
Comparison of Gene Expression Profiles from the Chromothripsis Chromosome for SCLC-21H against Intact Chromosomes, Related to Figure 7 Comparison of gene expression profiles from the chromothripsis chromosome (chr 8, blue line) for SCLC-21H against intact chromosomes (chr 5, 6 and 7; green, yellow and red lines respectively).The expression level of each gene is expressed as a Z score compared against the set of other SCLC cell lines (x axis), and it is the density / distribution of these Z scores that is plotted (y axis).
Comment in
- Cancer genomes evolve by pulverizing single chromosomes.
Meyerson M, Pellman D. Meyerson M, et al. Cell. 2011 Jan 7;144(1):9-10. doi: 10.1016/j.cell.2010.12.025. Cell. 2011. PMID: 21215363
Similar articles
- Mechanistic origins of diverse genome rearrangements in cancer.
Dahiya R, Hu Q, Ly P. Dahiya R, et al. Semin Cell Dev Biol. 2022 Mar;123:100-109. doi: 10.1016/j.semcdb.2021.03.003. Epub 2021 Apr 3. Semin Cell Dev Biol. 2022. PMID: 33824062 Free PMC article. Review. - Constitutional chromoanasynthesis: description of a rare chromosomal event in a patient.
Plaisancié J, Kleinfinger P, Cances C, Bazin A, Julia S, Trost D, Lohmann L, Vigouroux A. Plaisancié J, et al. Eur J Med Genet. 2014 Oct;57(10):567-70. doi: 10.1016/j.ejmg.2014.07.004. Epub 2014 Aug 13. Eur J Med Genet. 2014. PMID: 25128687 - In Brief: Chromothripsis and cancer.
Wyatt AW, Collins CC. Wyatt AW, et al. J Pathol. 2013 Sep;231(1):1-3. doi: 10.1002/path.4220. J Pathol. 2013. PMID: 23744564 - The Diverse Effects of Complex Chromosome Rearrangements and Chromothripsis in Cancer Development.
de Pagter MS, Kloosterman WP. de Pagter MS, et al. Recent Results Cancer Res. 2015;200:165-93. doi: 10.1007/978-3-319-20291-4_8. Recent Results Cancer Res. 2015. PMID: 26376877 Review. - Chromosomal catastrophe is a frequent event in clinically insignificant prostate cancer.
Kovtun IV, Murphy SJ, Johnson SH, Cheville JC, Vasmatzis G. Kovtun IV, et al. Oncotarget. 2015 Oct 6;6(30):29087-96. doi: 10.18632/oncotarget.4900. Oncotarget. 2015. PMID: 26337081 Free PMC article.
Cited by
- Sporadic and reversible chromothripsis in chronic lymphocytic leukemia revealed by longitudinal genomic analysis.
Bassaganyas L, Beà S, Escaramís G, Tornador C, Salaverria I, Zapata L, Drechsel O, Ferreira PG, Rodriguez-Santiago B, Tubio JM, Navarro A, Martín-García D, López C, Martínez-Trillos A, López-Guillermo A, Gut M, Ossowski S, López-Otín C, Campo E, Estivill X. Bassaganyas L, et al. Leukemia. 2013 Dec;27(12):2376-9. doi: 10.1038/leu.2013.127. Epub 2013 Apr 24. Leukemia. 2013. PMID: 23612016 Free PMC article. No abstract available. - Gene fusions by chromothripsis of chromosome 5q in the VCaP prostate cancer cell line.
Teles Alves I, Hiltemann S, Hartjes T, van der Spek P, Stubbs A, Trapman J, Jenster G. Teles Alves I, et al. Hum Genet. 2013 Jun;132(6):709-13. doi: 10.1007/s00439-013-1308-1. Epub 2013 Apr 25. Hum Genet. 2013. PMID: 23615946 - The origin and impact of embryonic aneuploidy.
Fragouli E, Alfarawati S, Spath K, Jaroudi S, Sarasa J, Enciso M, Wells D. Fragouli E, et al. Hum Genet. 2013 Sep;132(9):1001-13. doi: 10.1007/s00439-013-1309-0. Epub 2013 Apr 26. Hum Genet. 2013. PMID: 23620267 - Concepts in solid tumor evolution.
Sidow A, Spies N. Sidow A, et al. Trends Genet. 2015 Apr;31(4):208-14. doi: 10.1016/j.tig.2015.02.001. Epub 2015 Feb 27. Trends Genet. 2015. PMID: 25733351 Free PMC article. Review. - Getting personalized cancer genome analysis into the clinic: the challenges in bioinformatics.
Valencia A, Hidalgo M. Valencia A, et al. Genome Med. 2012 Jul 30;4(7):61. doi: 10.1186/gm362. eCollection 2012. Genome Med. 2012. PMID: 22839973 Free PMC article. Review.
References
- Akhoondi S., Sun D., von der Lehr N., Apostolidou S., Klotz K., Maljukova A., Cepeda D., Fiegl H., Dafou D., Marth C. FBXW7/hCDC4 is a general tumor suppressor in human cancer. Cancer Res. 2007;67:9006–9012. - PubMed
- Artandi S.E., Chang S., Lee S.L., Alson S., Gottlieb G.J., Chin L., DePinho R.A. Telomere dysfunction promotes non-reciprocal translocations and epithelial cancers in mice. Nature. 2000;406:641–645. - PubMed
- Bardeesy N., DePinho R.A. Pancreatic cancer biology and genetics. Nat. Rev. 2002;2:897–909. - PubMed
Publication types
MeSH terms
Grants and funding
- 088340/WT_/Wellcome Trust/United Kingdom
- 093867/WT_/Wellcome Trust/United Kingdom
- 077012/Z/05/Z/WT_/Wellcome Trust/United Kingdom
- WT088340MA/WT_/Wellcome Trust/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases