Enteric pathogen exploitation of the microbiota-generated nutrient environment of the gut - PubMed (original) (raw)
Review
Enteric pathogen exploitation of the microbiota-generated nutrient environment of the gut
Kristie M Keeney et al. Curr Opin Microbiol. 2011 Feb.
Abstract
Residing within the intestine is a large community of commensal organisms collectively termed the microbiota. This community generates a complex nutrient environment by breaking down indigestible food products into metabolites that are used by both the host and the microbiota. Both the invading intestinal pathogen and the microbiota compete for these metabolites, which can shape both the composition of the flora, as well as susceptibility to infection. After infection is established, pathogen mediated inflammation alters the composition of the microbiota, which further shifts the makeup of metabolites in the gastrointestinal tract. A greater understanding of the interplay between the microbiota, the metabolites they generate, and susceptibility to enteric disease will enable the discovery of novel therapies against infectious disease.
Copyright © 2010 Elsevier Ltd. All rights reserved.
Figures
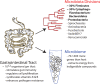
Figure 1. Functions of the host microbiota
Within the gastrointestinal (GI) tract is a community of commensal organisms, the microbiota, with an estimated density as high as 1012 organisms per gram of content [1]. Out of a total of 55 bacterial divisions identified thus far, only 8 have been identified within the human GI tract (dominant divisions are in bold) [2]. The genes encoded by this massive community are collectively termed the microbiome, which encodes an estimated 70–140 times more genes than encoded by its the human host [3,4]. Together, the organisms that reside in the GI tract and the genes they encode are necessary for the completion of essential tasks for the host, including stimulating gut immunity, regulating cell proliferation, vitamin synthesis, and mediating resistance to pathogen invasion and colonization.
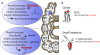
Figure 2. Chemical sensing between the microbiota and EHEC
(A) Members of the Bacteroidetes phyla produce acyl-homoserine lactones (AHLs). These signaling molecules are prominent within the rumen of the bovine gut, but not in other areas of the GI tract. AHLs isolated from the rumen stabilize folding of SdiA, an EHEC regulator that is necessary for colonization of cattle. Specifically, the rumen AHL-SdiA complex represses transcription and protein production by the locus of enterocyte effacement (LEE), a pathogenicity island that enables EHEC to colonize and promote disease in its human host, an undesirable phenotype for commensal colonization of cattle. Conversely, the AHL-SdiA complex activated the expression of gad acid-resistance genes and promoted survival in low pH, a phenotype necessary for EHEC survival within the acidic stomachs of the cow [36]. (B) Shiga Toxin 2 (Stx2) is a major virulence factor of EHEC O157:H7, which causes protein synthesis inhibition and ultimately cell death in the human host. Prokaryotes of conventionalized rats colonized with human microbiota produced molecules which repressed RecA mediated stx2 mRNA expression and Stx2 production. Subsequent analysis revealed that these inhibitory prokaryotic molecules are produced in part by Bacteroides thetaiotaomicron, a member of the normal human intestinal microbiota [37].
Figure 3. Short-chain fatty acid (SCFA) influence upon pathogen tropism
(A) The Firmicutes are a principal phyla in both the small intestine and the colon, with the family Lachnospiraceae dominating the colon [21,53]. The Lachnospiraceae are members of the Clostridia class, which are major producers of butyrate in the human colon [43,53]. The Lactobacillales order of the Bacillus class dominate the small intestine in humans, and upon further examination in mice, the family Lactobacillaceae within this order compose 24% of the total small intestine microbiota [21,53]. Genera belonging to this family include Lactobacillus, which heterofermentatively can produce formate as well as acetate and lactate. (B) EHEC primarily colonizes the colon of humans, where butyrate is a dominant SCFA [21,39,44,45]. In EHEC, butyrate activates the locus of enterocyte effacement (LEE) and enhances adherence of this pathogen in tissue culture [39,48]. Salmonella enterica serovar Typhimurium (S. Typhimurium) colonizes the small intestine, where formate is a dominant SCFA. The SCFA formate induces the expression of invasion genes in S. Typhimurium, while butyrate is known to repress these genes [46,47].
Similar articles
- Gut microbiota and its pathophysiology in disease paradigms.
Festi D, Schiumerini R, Birtolo C, Marzi L, Montrone L, Scaioli E, Di Biase AR, Colecchia A. Festi D, et al. Dig Dis. 2011;29(6):518-24. doi: 10.1159/000332975. Epub 2011 Dec 12. Dig Dis. 2011. PMID: 22179206 Review. - When pathogenic bacteria meet the intestinal microbiota.
Rolhion N, Chassaing B. Rolhion N, et al. Philos Trans R Soc Lond B Biol Sci. 2016 Nov 5;371(1707):20150504. doi: 10.1098/rstb.2015.0504. Philos Trans R Soc Lond B Biol Sci. 2016. PMID: 27672153 Free PMC article. Review. - The role of the gut microbiota in nutrition and health.
Flint HJ, Scott KP, Louis P, Duncan SH. Flint HJ, et al. Nat Rev Gastroenterol Hepatol. 2012 Sep 4;9(10):577-89. doi: 10.1038/nrgastro.2012.156. eCollection 2012 Oct. Nat Rev Gastroenterol Hepatol. 2012. PMID: 22945443 Review. - Influence of fermented milk products, prebiotics and probiotics on microbiota composition and health.
Ceapa C, Wopereis H, Rezaïki L, Kleerebezem M, Knol J, Oozeer R. Ceapa C, et al. Best Pract Res Clin Gastroenterol. 2013 Feb;27(1):139-55. doi: 10.1016/j.bpg.2013.04.004. Best Pract Res Clin Gastroenterol. 2013. PMID: 23768559 Review. - Nutrition, the gut microbiome and the metabolic syndrome.
Kovatcheva-Datchary P, Arora T. Kovatcheva-Datchary P, et al. Best Pract Res Clin Gastroenterol. 2013 Feb;27(1):59-72. doi: 10.1016/j.bpg.2013.03.017. Best Pract Res Clin Gastroenterol. 2013. PMID: 23768553 Review.
Cited by
- Enterohaemorrhagic E. coli utilizes host- and microbiota-derived L-malate as a signaling molecule for intestinal colonization.
Liu B, Jiang L, Liu Y, Sun H, Yan J, Kang C, Yang B. Liu B, et al. Nat Commun. 2023 Nov 9;14(1):7227. doi: 10.1038/s41467-023-43149-7. Nat Commun. 2023. PMID: 37945607 Free PMC article. - A Nitrogen Metabolic Enzyme Provides Salmonella Fitness Advantage by Promoting Utilization of Microbiota-Derived Carbon Source.
Yoo W, Choi J, Park B, Byndloss MX, Ryu S. Yoo W, et al. ACS Infect Dis. 2021 May 14;7(5):1208-1220. doi: 10.1021/acsinfecdis.0c00836. Epub 2021 Apr 14. ACS Infect Dis. 2021. PMID: 33853321 Free PMC article. - How microbiomes can help inform conservation: landscape characterisation of gut microbiota helps shed light on additional population structure in a specialist folivore.
Littleford-Colquhoun BL, Weyrich LS, Hohwieler K, Cristescu R, Frère CH. Littleford-Colquhoun BL, et al. Anim Microbiome. 2022 Jan 31;4(1):12. doi: 10.1186/s42523-021-00122-3. Anim Microbiome. 2022. PMID: 35101152 Free PMC article. - Campylobacter jejuni pdxA affects flagellum-mediated motility to alter host colonization.
Asakura H, Hashii N, Uema M, Kawasaki N, Sugita-Konishi Y, Igimi S, Yamamoto S. Asakura H, et al. PLoS One. 2013 Aug 6;8(8):e70418. doi: 10.1371/journal.pone.0070418. Print 2013. PLoS One. 2013. PMID: 23936426 Free PMC article. - Role of the gut microbiota in nutrient competition and protection against intestinal pathogen colonization.
Horrocks V, King OG, Yip AYG, Marques IM, McDonald JAK. Horrocks V, et al. Microbiology (Reading). 2023 Aug;169(8):001377. doi: 10.1099/mic.0.001377. Microbiology (Reading). 2023. PMID: 37540126 Free PMC article. Review.
References
- Bäckhed F, Ley RE, Sonnenburg JL, Peterson DA, Gordon JI. Host-bacterial mutualism in the human intestine. Science. 2005;307:1915–1920. - PubMed
- Turnbaugh PJ, Hamady M, Yatsunenko T, Cantarel BL, Duncan A, Ley RE, Sogin ML, Jones WJ, Roe BA, Affourtit JP, et al. A core gut microbiome in obese and lean twins. Nature. 2009;457:480–484. - PMC - PubMed
- In this study, the authors characterized the microbiota and microbiomes of lean and obese twin pairs. They demonstrated that a core microbiome of shared genes exists between all individuals they characterized, and that between the obese and lean pairs, shifts in the composition of the microbiota correlated with corresponding shifts in microbiome genes that are involved in energy uptake.
- Possemiers S, Grootaert C, Vermeiren J, Gross G, Marzorati M, Verstraete W, Van de Wiele T. The intestinal environment in health and disease - recent insights on the potential of intestinal bacteria to influence human health. Curr Pharm Des. 2009;15:2051–2065. - PubMed
- Zhang H, DiBaise JK, Zuccolo A, Kudrna D, Braidotti M, Yu Y, Parameswaran P, Crowell MD, Wing R, Rittmann BE, et al. Human gut microbiota in obesity and after gastric bypass. Proc Natl Acad Sci USA. 2009;106:2365–2370. - PMC - PubMed
- The authors demonstrate by 454 pyrosequencing of 3 normal weight, 3 morbidly obese, and 3 post-gastric-bypass surgery individuals that some members of the microbiota are more abundant in the obese. Specifically, they saw increased numbers of H(2)-producing Prevotellaceae and H(2)-utilizing methanogenic Archaea in obese individuals, and they hypothesized that interpecies H(2)-transfer between these populations may be an important mechanism to increase energy uptake in the obese.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources